Abstract
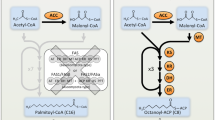
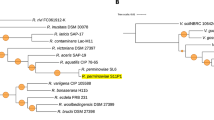
Arbuscular mycorrhizal (AM) fungi are ubiquitous endosymbionts of terrestrial plants. It helps plants to extract more nutrients from the soil and enhances the plant tolerance to various ecological stress factors. The AM fungal genome sequence helps to identify the gene repertoires that are crucial for adaptation to different habitat and mechanisms for interaction with host plant. The present work comprises the first draft of the genome sequence of Rhizophagus proliferus, which is an important AM species present in biofertilizer consortia for agricultural purpose. The estimated genome size of R. proliferus is ~ 110 Mbps and its genomic assembly is 94.35% complete. Genome mining was carried out to identify putative gene families important for biological functions. A total of 22,526 protein-coding genes were estimated in the genome, with an abundance of kinases and reduced number of glycoside hydrolases as compared to other fungal classes. A striking finding in the R. proliferus genome was higher number of carbohydrate esterases (CE), which may suggest towards presence of higher saprotrophic activity in this species as compared to the previously reported AM fungi, which may indicate towards its role as a link between plants and soil mineral nutrients. The genome sequence and annotation of R. proliferus presented here would serve as an important reference for functional genomics studies required for developing biofertilizer formulations in future. In addition, the findings from this work may also prove important in deciphering molecular mechanisms in AM fungi that govern the host-specific interaction and associated agriculture benefits.




Similar content being viewed by others
Data availability
The whole genome project has been deposited at https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA392539 under Bioproject number PRJNA392539. The R. proliferus Internal Transcribed Spacer (ITS) gene sequence is deposited at Genbank with Accession number OK030706. All data generated and analysed during this study are included in this report and its supplementary information files.
Abbreviations
- CMCC:
-
Centre for mycorrhizal culture collection
- HMW:
-
High molecular weight
- NCBI:
-
National center for biotechnology information
- KOG:
-
Eukaryotic orthologous groups
- CAZyme:
-
Carbohydrate active enzyme
- BLAST:
-
Basic local alignment search tool
- GPCRs:
-
G-protein-coupled receptors
- Blastp:
-
Protein BLAST: search protein databases using a protein query
- CDD:
-
NCBI conserved domain database
- SMART:
-
Simple modular architecture research tool database
- AA:
-
Auxiliary activity
- CBM:
-
Carbohydrate-binding modules
- CE:
-
Carbohydrate esterases
- EXPN:
-
Expansins
- GH:
-
Glycoside hydrolases
- GT:
-
Glycosyltransferases
- PL:
-
Polysaccharide lyases
- DMAT:
-
DiMethylAllyl tryptophan synthase
- NRPS:
-
Non-ribosomal peptide synthetases
- PKS:
-
PolyKetide synthetases
- TS:
-
Terpene synthases
References
Błaszkowski J, Chwat G, Góralska A, Ryszka P, Orfanoudakis M (2014) Septoglomus jasnowskae and Septoglomus turnauae, two new species of arbuscular mycorrhizal fungi (Glomeromycota). Mycol Progress 13:999–1009. https://doi.org/10.1007/s11557-014-0985-z
Bravo A, Brands M, Wewer V, Dörmann P, Harrison MJ (2017) Arbuscular mycorrhiza-specific enzymes FatM and RAM2 fine-tune lipid biosynthesis to promote development of arbuscular mycorrhiza. New Phytol 214:1631–1645. https://doi.org/10.1111/nph.14533
Bunn RA, Simpson DT, Bullington LS, Lekberg Y, Janos D (2019) Revisiting the ‘direct mineral cycling’ hypothesis: arbuscular mycorrhizal fungi colonize leaf litter, but why? ISME J 13:1891–1898. https://doi.org/10.1038/s41396-019-0403-2
Casieri L, Ait Lahmidi N, Doidy J, Veneault-Fourrey C, Migeon A, Bonneau L, Courty P, Garcia K, Charbonnier M, Delteil A, Brun A, Zimmermann S, Plassard C, Wipf D (2013) Biotrophic transportome in mutualistic plant-fungal interactions. Mycorrhiza 23:597–625. https://doi.org/10.1007/s00572-013-0496-9
Cherry JM, Hong EL, Amundsen C, Balakrishnan R, Binkley G, Chan ET, Christie KR, Costanzo MC, Dwight SS, Engel SR, Fisk DG, Hirschman JE, Hitz BC, Karra K, Krieger CJ, Miyasato SR, Nash RS, Park J, Skrzypek MS, Simison M, Weng S, Wong ED (2012) Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res 40:D700–D705. https://doi.org/10.1093/nar/gkr1029
Cosgrove DJ, Li LC, Cho HT, Hoffmann-Benning S, Moore RC, Blecker D (2002) The growing world of expansins. Plant Cell Physiol 43:1436–1444. https://doi.org/10.1093/pcp/pcf180
Declerck S, Cranenbrouck S, Dalpé Y, Séguin S, Grandmougin-Ferjani A, Fontaine J, Sancholle M (2000) Glomus proliferum sp. nov.: A description based on morphological, biochemical. Molecular and Monoxenic Cultivation Data Mycologia 92:1178–1187. https://doi.org/10.1080/00275514.2000.12061265
Halary S, Malik SB, Lildhar L, Slamovits CH, Hijri M, Corradi N (2011) Conserved meiotic machinery in Glomus spp., a putatively ancient asexual fungal lineage. Genome Biol Evol 3:950–958. https://doi.org/10.1093/gbe/evr089
Jiang Y, Wang W, Xie Q, Liu N, Liu L, Wang D, Zhang X, Yang C, Chen X, Tang D, Wang E (2017) Plants transfer lipids to sustain colonization by mutualistic mycorrhizal and parasitic fungi. Science 356:1172–1175. https://doi.org/10.1126/science.aam9970
Jung SC, Martinez-Medina A, Lopez-Raez JA, Pozo MJ (2012) Mycorrhiza-Induced Resistance and Priming of Plant Defenses. J Chem Ecol 38:651–664. https://doi.org/10.1007/s10886-012-0134-6
Keymer A, Huber C, Eisenreich W, Gutjahr C (2018) Tracking Lipid Transfer by Fatty Acid Isotopolog Profiling from Host Plants to Arbuscular Mycorrhiza Fungi. BIO-PROTOCOL 8:e2786. https://doi.org/10.21769/BioProtoc.2786
Kobayashi Y, Maeda T, Yamaguchi K, Kameoka H, Tanaka S, Ezawa T, Shigenobu S, Kawaguchi M (2018) The genome of Rhizophagus clarus HR1 reveals a common genetic basis for auxotrophy among arbuscular mycorrhizal fungi. BMC Genomics 19(1):465. https://doi.org/10.1186/s12864-018-4853-0
Kristiina H, Miia Mäkelä R (2018) Role of fungi in wood decay. Reference Module Life Sciences Elsevier. https://doi.org/10.1016/B978-0-12-809633-8.12424-0
Krüger M, Krüger C, Walker C, Stockinger H, Schüßler A (2012) Phylogenetic reference data for systematics and phylotaxonomy of arbuscular mycorrhizal fungi from phylum to species level. New Phytol 193(4):970–984. https://doi.org/10.1111/j.1469-8137.2011.03962.x
Leibundgut M, Maier T, Jenni S, Ban N (2008) The multienzyme architecture of eukaryotic fatty acid synthases. Curr Opin Struct Biol 18:714–725. https://doi.org/10.1016/j.sbi.2008.09.008
Leyval C, Joner EJ, del Val C, Haselwandter K (2002) Potential of arbuscular mycorrhizal fungi for bioremediation. In: Gianinazzi S, Schüepp H, Barea JM, Haselwandter K (eds) Mycorrhizal Technology in Agriculture. Birkhäuser, Basel, pp 175–186. https://doi.org/10.1007/978-3-0348-8117-3_14
Lin K, Limpens E, Zhang Z, Ivanov S, Saunders DG, Mu D, Pang E, Cao H, Cha H, Lin T, Zhou Q, Shang Y, Li Y, Sharma T, van Velzen R, de Ruijter N, Aanen DK, Win J, Kamoun S, Bisseling T, Geurts R, Huang S (2014) Single nucleus genome sequencing reveals high similarity among nuclei of an endomycorrhizal fungus. PLoS Genet 10(1):e1004078. https://doi.org/10.1371/journal.pgen.1004078
Logi C, Sbrana C, Giovannetti M (1998) Cellular events involved in survival of individual arbuscular mycorrhizal symbionts growing in the absence of the host. Appl Environ Microbiol 64:3473–3479. https://doi.org/10.1128/AEM.64.9.3473-3479.1998
Ma LJ, Ibrahim AS, Skory C, Grabherr MG, Burger G, Butler M, Elias M, Idnurm A, Lang BF, Sone T, Abe A, Calvo SE, Corrochano LM, Engels R, Fu J, Hansberg W, Kim JM, Kodira CD, Koehrsen MJ, Liu B, Miranda-Saavedra D, O’Leary S, Ortiz-Castellanos L, Poulter R, Rodriguez-Romero J, Ruiz-Herrera J, Shen YQ, Zeng Q, Galagan J, Birren BW, Cuomo CA, Wickes BL (2009) Genomic analysis of the basal lineage fungus Rhizopus oryzae reveals a whole-genome duplication. PLoS Genet 5(7):e1000549. https://doi.org/10.1371/journal.pgen.1000549
Martin F, Aerts A, Ahrén D, Brun A, Danchin EG, Duchaussoy F, Gibon J, Kohler A, Lindquist E, Pereda V, Salamov A, Shapiro HJ, Wuyts J, Blaudez D, Buée M, Brokstein P, Canbäck B, Cohen D, Courty PE, Coutinho PM, Delaruelle C, Detter JC, Deveau A, DiFazio S, Duplessis S, Fraissinet-Tachet L, Lucic E, Frey-Klett P, Fourrey C, Feussner I, Gay G, Grimwood J, Hoegger PJ, Jain P, Kilaru S, Labbé J, Lin YC, Legué V, Le Tacon F, Marmeisse R, Melayah D, Montanini B, Muratet M, Nehls U, Niculita-Hirzel H, Oudot-Le Secq MP, Peter M, Quesneville H, Rajashekar B, Reich M, Rouhier N, Schmutz J, Yin T, Chalot M, Henrissat B, Kües U, Lucas S, Van de Peer Y, Podila GK, Polle A, Pukkila PJ, Richardson PM, Rouzé P, Sanders IR, Stajich JE, Tunlid A, Tuskan G, Grigoriev IV (2008) The genome of Laccaria bicolor provides insights into mycorrhizal symbiosis. Nature 452:88–92. https://doi.org/10.1038/nature06556
Martin F, Kohler A, Murat C, Balestrini R, Coutinho PM, Jaillon O, Montanini B, Morin E, Noel B, Percudani R, Porcel B, Rubini A, Amicucci A, Amselem J, Anthouard V, Arcioni S, Artiguenave F, Aury JM, Ballario P, Bolchi A, Brenna A, Brun A, Buée M, Cantarel B, Chevalier G, Couloux A, Da Silva C, Denoeud F, Duplessis S, Ghignone S, Hilselberger B, Iotti M, Marçais B, Mello A, Miranda M, Pacioni G, Quesneville H, Riccioni C, Ruotolo R, Splivallo R, Stocchi V, Tisserant E, Viscomi AR, Zambonelli A, Zampieri E, Henrissat B, Lebrun MH, Paolocci F, Bonfante P, Ottonello S, Wincker P (2010) Périgord black truffle genome uncovers evolutionary origins and mechanisms of symbiosis. Nature 464:1033–1038. https://doi.org/10.1038/nature08867
Melo CD, Luna S, Krüger C, Walker C, Mendonça D, Fonseca MACH, Jaizme-Vega M, Machado CA (2018) Communities of arbuscular mycorrhizal fungi under Picconia azorica in native forests of Azores. Symbiosis 74:43–54. https://doi.org/10.1007/s13199-017-0487-2
Morin E, Miyauchi S, San Clemente H, Chen ECH, Pelin A, de la Providencia I, Ndikumana S, Beaudet D, Hainaut M, Drula E, Kuo A, Tang N, Roy S, Viala J, Henrissat B, Grigoriev IV, Corradi N, Roux C, Martin FM (2019) Comparative genomics of Rhizophagus irregularis, R. cerebriforme, R. diaphanus and Gigaspora rosea highlights specific genetic features in Glomeromycotina. New Phytol 222:1584–1598. https://doi.org/10.1111/nph.15687
Morley KL, Chauve G, KazlauskasvR DC, Shareck F, Marchessault RH (2006) Acetyl xylan esterase-catalyzed deacetylation of chitin and chitosan. Carbohyd Polym 63(3):310–315. https://doi.org/10.1016/j.carbpol.2005.07.034
Peña Venegas RA, Lee SJ, Thuita M, Mlay DP, Masso C, Vanlauwe B, Rodriguez A, Sanders IR (2021) The phosphate inhibition paradigm: host and fungal genotypes determine arbuscular mycorrhizal fungal colonization and responsiveness to inoculation in cassava with increasing phosphorus supply. Front Plant Sci 12:693037. https://doi.org/10.3389/fpls.2021.693037
Prasad Singh P, Srivastava D, Jaiswar A, Adholeya A (2019) Effector proteins of Rhizophagus proliferus: conserved protein domains may play a role in host-specific interaction with different plant species. Braz J Microbiol 50(3):593–601. https://doi.org/10.1007/s42770-019-00099-x
Riley R, Charron P, Idnurm A, Farinelli L, Dalpé Y, Martin F, Corradi N (2014) Extreme diversification of the mating type-high-mobility group (MATA-HMG) gene family in a plant-associated arbuscular mycorrhizal fungus. New Phytol 201:254–268. https://doi.org/10.1111/nph.12462
Ropars J, Toro KS, Noel J, Pelin A, Charron P, Farinelli L, Marton T, Krüger M, Fuchs J, Brachmann A, Corradi N (2016) Evidence for the sexual origin of heterokaryosis in arbuscular mycorrhizal fungi. Nat Microbiol 1(6):16033. https://doi.org/10.1038/nmicrobiol.2016.33
Salvioli A, Ghignone S, Novero M, Navazio L, Venice F, Bagnaresi P, Bonfante P (2016) Symbiosis with an endobacterium increases the fitness of a mycorrhizal fungus, raising its bioenergetic potential. ISME J 10:130–144. https://doi.org/10.1038/ismej.2015.91
Sanders IR, Croll D (2010) Arbuscular mycorrhiza: the challenge to understand the genetics of the fungal partner. Annu Rev Genet 44:271–292. https://doi.org/10.1146/annurev-genet-102108-134239
Schüßler A, Sschwarzott D, Walker C (2001) A new fungal phylum, the Glomeromycota: phylogeny and evolution. Mycol Res 105:1413–1421. https://doi.org/10.1017/S0953756201005196
Schütz L, Gattinger A, Meier M, Müller A, Boller T, Mäder P, Mathimaran N (2018) Improving crop yield and nutrient use efficiency via biofertilization—a global meta-analysis. Front Plant Sci 8:2204. https://doi.org/10.3389/fpls.2017.02204
Shachar-Hill Y, Pfeffer PE, Douds D, Osman SF, Doner LW, Ratcliffe RG (1995) Partitioning of intermediary carbon metabolism in vesicular-arbuscular mycorrhizal leek. Plant Physiol 108:7–15. https://doi.org/10.1104/pp.108.1.7
Shen M, Zhao DK, Qiao Q, Liu L, Wang JL, Cao GH, Li T, Zhao ZW (2015) Identification of glutathione S-transferase (GST) genes from a dark septate endophytic fungus (Exophiala pisciphila) and their expression patterns under varied metals stress. PLoS ONE 10(4):e0123418. https://doi.org/10.1371/journal.pone.0123418
Sista Kameshwar AK, Qin W (2018) Understanding the structural and functional properties of carbohydrate esterases with a special focus on hemicellulose deacetylating acetyl xylan esterases. Mycology 9(4):273–295. https://doi.org/10.1080/21501203.2018.1492979
Smith SE, Read D (2008) Mycorrhizal symbiosis, 3rd edn. Academic press, Australia, pp 611–636
Sokolski S, Dalpé Y, Seguin S, Khasa D, Lévesque C, Piché Y (2010) Conspecificity of DAOM 197198, the model arbuscular mycorrhizal fungus, with Glomus irregulare: molecular evidence with three protein-encoding genes. Botany 88:829–838. https://doi.org/10.1139/B10-050
Spanu PD, Abbott JC, Amselem J et al (2010) Genome expansion and gene loss in powdery mildew fungi reveal tradeoffs in extreme parasitism. Science 330(6010):1543–1546. https://doi.org/10.1126/science.1194573
Stockinger H, Krüger M, Schüßler A (2010) DNA barcoding of arbuscular mycorrhizal fungi. New Phytol 187:461–474. https://doi.org/10.1111/j.1469-8137.2010.03262.x
Sun X, Chen W, Ivanov S, MacLean AM, Wight H, Ramaraj T, Mudge J, Harrison MJ, Fei Z (2018) Genome and evolution of the arbuscular mycorrhizal fungus Diversispora epigaea (formerly Glomus versiforme) and its bacterial endosymbionts. New Phytol 221:1556–1573. https://doi.org/10.1111/nph.15472
Sun X, Zhu J, Bao L, Hu C, Jin C, Harris SD, Liu H, Li S (2013) PyrG is required for maintaining stable cellular uracil level and normal sporulation pattern under excess uracil stress in Aspergillus nidulans. Sci China Life Sci 56:467–475. https://doi.org/10.1007/s11427-013-4480-6
Tang N, San Clemente H, Roy S, Bécard G, Zhao B, Roux C (2016) A Survey of the Gene Repertoire of Gigaspora rosea Unravels Conserved Features among Glomeromycota for Obligate Biotrophy. Front Microbiol 7:233. https://doi.org/10.3389/fmicb.2016.00233
Tisserant E, Kohler A, Dozolme-Seddas P (2012) The transcriptome of the arbuscular mycorrhizal fungus Glomus intraradices (DAOM 197198) reveals functional tradeoffs in an obligate symbiont. New Phytol 193(3):755–769. https://doi.org/10.1111/j.1469-8137.2011.03948.x
Tisserant E, Malbreil M, Kuo A, Kohler A, Symeonidi A, Balestrini R, Charron P, Duensing N, Frei dit Frey N, Gianinazzi-Pearson V, Gilbert LB, Handa Y, Herr JR, Hijri M, Koul R, Kawaguchi M, Krajinski F, Lammers PJ, Masclaux FG, Murat C, Morin E, Ndikumana S, Pagni M, Petitpierre D, Requena N, Rosikiewicz P, Riley R, Saito K, San Clemente H, Shapiro H, van Tuinen D, Bécard G, Bonfante P, Paszkowski U, Shachar-Hill YY, Tuskan GA, Young JP, Sanders IR, Henrissat B, Rensing SA, Grigoriev IV, Corradi N, Roux C, Martin F (2013) Genome of an arbuscular mycorrhizal fungus provides insight into the oldest plant symbiosis. Proceedings of the National Academy of Sciences of the United States of America 110:20117–20122. https://doi.org/10.1073/pnas.1313452110
Treseder KK, Allen EB, Egerton-Warburton LM, Hart MM, Klironomos JN, Maherali H, Tedersoo L (2018) Arbuscular mycorrhizal fungi as mediators of ecosystem responses to nitrogen deposition: a trait-based predictive framework. J Ecol 106(2):480–489. https://doi.org/10.1111/1365-2745.12919
Van der Heijden MAG, Klironomos JN, Ursic M, Moutoglis P, Streitwolf-Engel R, Boller T, Wiemken A, Sanders IR (1998) Mycorrhizal fungal diversity determines plant biodiversity, ecosystem variability and productivity. Nature 396:69–72. https://doi.org/10.1038/23932
Venice F, Ghignone S, Salvioli di Fossalunga A, Amselem J, Novero M, Xianan X, Sędzielewska Toro K, Morin E, Lipzen A, Grigoriev IV, Henrissat B, Martin FM, Bonfante P (2020) At the nexus of three kingdoms: the genome of the mycorrhizal fungus Gigaspora margarita provides insights into plant, endobacterial and fungal interactions. Environ Microbiol 22:122–141. https://doi.org/10.1111/1462-2920.14827
Voß S, Betz R, Heidt S, Corradi N, Requena N (2018) RiCRN1, a Crinkler Effector From the Arbuscular Mycorrhizal Fungus Rhizophagus irregularis, Functions in Arbuscule Development. Front Microbiol 9:2068. https://doi.org/10.3389/fmicb.2018.02068
Yang Y, Liang Y, Han X, Chiu T, Ghosh A, Chen H, Tang M (2016) The roles of arbuscular mycorrhizal fungi (AMF) in phytoremediation and tree-herb interactions in Pb contaminated soil. Sci Rep 6:20469. https://doi.org/10.1038/srep20469
Yelamanchi SD, Jayaram S, Thomas JK, Gundimeda S, Khan AA, Singhal A, Keshava Prasad TS, Pandey A, Somani BL, Gowda H (2016) A pathway map of glutamate metabolism. J Cell Commun Signal 10(1):69–75. https://doi.org/10.1007/s12079-015-0315-5
Acknowledgements
We are thankful to Ms. Pratima Vasistha for supporting by providing spores from monosporal culture of R. proliferus and carrying out morphological analysis. We thank Dr. Alok Adholeya for initial discussions regarding the project idea.
Funding
This work was supported by the grant-in-aid for research to Dr. Pushplata Prasad Singh by the Science and Engineering Research Board (SERB), Department of Science and Technology (DST), India, under the grant number “EMR/2017/000657”. Funders had no role in the study.
Author information
Authors and Affiliations
Contributions
All authors have read and approved the final manuscript. PPS was involved in conceptualization of the project, study design, data analysis, data compilation, manuscript writing, critical inputs and finalization of the manuscript. DS was involved in genomics data analysis, data compilation and manuscript writing. SS carried out DNA extraction and molecular characterization. V was involved in raising monosporal cultures and morphology-based characterization of spores.
Corresponding author
Ethics declarations
Conflict of interests
None of the authors declare any competing interests.
Ethics approval
Not Applicable.
Consent to participate
Not Applicable.
Consent for publication
Not Applicable.
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Singh, P.P., Srivastava, D., Shukla, S. et al. Rhizophagus proliferus genome sequence reiterates conservation of genetic traits in AM fungi, but predicts higher saprotrophic activity. Arch Microbiol 204, 105 (2022). https://doi.org/10.1007/s00203-021-02651-6
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00203-021-02651-6