Abstract
Strain CAP181T, an endophytic actinobacterium, was isolated from a surface sterilized root sample of a native pine tree, Flinders University, Adelaide, South Australia. Chemotaxonomic data including cell wall components, major fatty acids, and major menaquinones confirmed the affiliation of strain CAP181T to the genus Micromonospora. This strain was Gram stain positive with well-developed substrate mycelia to form a single spore with hairy surface. The phylogenetic tree showed that M. coerulea NBRC 13504 T is the closest phylogenetic neighbour, sharing 99.2% 16S rRNA gene similarity and the next closest neighbor is M. chaiyaphumensis DSM 45246 T (98.7%). Genome mining of this strain revealed genes encoding to enzymes relating to nitrogen fixation and bioremediation. Based on genotypic and phenotypic studies including DNA–DNA hybridization data, strain CAP181T was different from any of the closely related species with valid names. The name proposed for the new species is Micromonospora veneta sp. nov. The type strain is CAP181T (= DSM 109713 T = NRRL B-65535 T).


Similar content being viewed by others
References
Atlas RM, Parks LC (1993) In: Parks LC (ed) Handbook of microbiological media. CRC Press, Boca Raton
Baldani JI, Caruso L, Baldani VLD, Goi SR, Döbereiner J (1997) Recent advances in BNF with non-legume plants. Soil Biol Biochem 29:911–922
Blin K, Shaw S, Steinke K, Villebro R, Ziemert N, Lee SY, Medema MH, Weber T (2019) AntiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res 47:W81–W87
Bousfield IJ, Keddie RM, Dando TR, Shaw S (1985) Simple rapid methods of cell wall analysis as an aid in the identification of aerobic coryneform bacteria. Chem Method Bact Syst Tech Ser 20:221–236
Carro L, Pukall R, Spröer C, Kroppenstedt RM, Trujillo ME (2012) Micromonospora cremea sp. nov. and Micromonospora zamorensis sp. nov., isolated from the rhizosphere of Pisum sativum. Int J Syst Evol Microbiol 62:2971–2977
Carro L, Riesco R, Spröer C, Trujillo ME (2016) Micromonospora luteifusca sp. nov. isolated from cultivated Pisum sativum. Syst Appl Microbiol 39:237–242
Carro L, Veyisoglu A, Riesco R, Spröer C, Klenk HP, Sahin N, Trujillo ME (2018) Micromonospora phytophila sp. nov. and Micromonospora luteiviridis sp. nov., isolated as natural inhabitants of plant nodules. Int J Syst Evol Microbiol 68:248–253
Chun J, Oren A, Ventosa A, Christensen H, Arahal DR, da Costa MS, Rooney AP, Yi H, Xu X-W, De Meyer S, Trujillo ME (2018) Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol 68:461–466
Ezaki T, Hashimoto Y, Yabuuchi E (1989) Fluorometric deoxyribonucleic acid-deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int J Syst Bacteriol 39:224–229
Farris JS (1972) Estimating phylogenetic trees from distance matrices. Am Nat 106:645–667
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Garcia LC, Martínez ME, Trujillo ME (2010) Micromonospora pisi sp. nov., isolated from root nodules of Pisum sativum. Int J Syst Evol Microbiol 60:331–337
Gordon RE, Barnett DA, Handerhan JE, Pang CH (1974) Nocardia coeliaca, Nocardia autotrophica, and the nocardin strain. Int J Syst Bacteriol 24:54–63
Genilloud O (2012) Genus I. Micromonospora Ørskov 1923, 156AL p. 1039–1057. In: Whitman WB, Goodfellow M, Kämpfer P, Busse HJ, Trujillo ME, Ludwig W, Suzuki KI, Parte A (eds) Bergey’s manual of systematic bacteriology, vol 4, 2nd edn. Springer, New York, p 1750
Hasegawa T, Takisawa M, Tanida S (1983) A rapid analysis for chemical grouping of aerobic actinomycetes. J Gen Appl Microbiol 29:319–322
Kaewkla O, Franco CMM (2013) Rational approaches to improving the isolation of endophytic actinobacteria from Australian native trees. Microbial Ecol 65:384–393
Kaewkla O, Franco CMM (2019) Actinomycetospora callitridis sp. nov., an endophytic actinobacterium isolated from the surface-sterilised root of an Australian native pine tree. Antonie Van Leeuwenhoek 112:331–337
Kang HS, Brady SF (2014) Arixanthomycins A−C: phylogeny-guided discovery of biologically active eDNA-derived pentangular polyphenols. ACS Chem Biol 9:1267–1272
Komagata K, Suzuki K (1987) Lipid and cell wall analysis in bacterial systematics. Methods Microbiol 19:161–207
Kornerup A, Wanscher JH (1978) Methuen handbook of colour, 3rd edn. Introduced and rev. by Pavey D. Eyre Methuen, London
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549
Kurup KV, Schmitt JA (1973) Numerical taxonomy of Nocardia. Can J Microbiol 19:1035–1048
Kusunoki S, Ezaki T, Tamesada M, Hatanaka Y, Asano K et al (1991) Application of colorimetric microdilution plate hybridization for rapid genetic identification of 22 mycobacterium species. J Clinic Microbiol 29:1596–1603
Lechevalier MP, Bievre CD, Lechevalier H (1977) Chemotaxonomy of aerobic actinomycetes: phospholipid composition. Biochem Syst Ecol 5:249–260
Lefort V, Desper R, Gascuel O (2015) FastME 2.0: a comprehensive, accurate, and fast distance-based phylogeny inference program. Mol Biol Evol 32:2798–2800
Meier-Kolthoff JP, Auch AF, Klenk H-P, Goker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. https://doi.org/10.1186/1471-2105-14-60
Meier-Kolthoff JP, Göker M (2019) TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat Commun 10:2182. https://doi.org/10.1038/s41467-019-
Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M, other authors, (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241
Nei M, Kumar S (2000) Molecular evolution and phylogenetics. Oxford University Press, New York
Parte AC, Sardà Carbasse J, Meier-Kolthoff JP, Reimer LC, Göker M (2020) List of Prokaryotic names with Standing in Nomenclature (LPSN) moves to the DSMZ. Int J Syst Evol Microbiol Microbiol 70:5607–5612
Richter M, Rosselló-Móra R (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 106:19126–19131
Richter M, Rosselló-Móra R, Oliver Glöckner F, Peplies J (2016) JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32:929–931
Sasser M (2001) Identification of bacteria by gas chromatography of cellular fatty acids. Technical note# 101 (http: www.midi-inc.com)
Schoenborn L, Yates PS, Grinton BE, Hugenholtz P, Janssen PH (2004) Liquid serial dilution is inferior to solid media for isolation of cultures representative of the phylum-level diversity of soil bacteria. Appl Environ Microbiol 70:4363–4366
Seemann T (2014) Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069
Shirling EB, Gottlieb D (1966) Methods for characterization of Micromonospora species. Int J Syst Bacteriol 16:313–340
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526
Talukdar M, Bora TC, Jha DK (2016) Micromonospora: A Potential Source of Antibiotic. In :Purkayastha P (eds). Bioprospecting of Indigenous Bioresources of North-East India; Singapore, Springer, 347 pp
Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J (2016) NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44:6614–6624
The UniProt Consortium (2019) UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res 47:D506-515
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Trujillo ME, Kroppenstedt RM, Schumann P, Carro L, Martínez Molina E (2006) Micromonospora coriariae sp. nov., isolated from root nodules of Coriaria myrtifolia. Int J Syst Evol Microbiol 56:2381–2385
Trujillo ME, Kroppenstedt RM, Fernádez-Molinero C, Schumann P, Martínez Molina E (2007) Micromonospora lupini sp. Nov. and Micromonospora saelicesensis sp. Nov., isolated from root nodules of Lupinus angustifolius. Int J Syst Evol Microbiol 57:2799–2804
Trujillo ME, Riesco R, Benito P, Carro L (2015) Endophytic actinobacteria and the interaction of Micromonospora and nitrogen fixing plants. Front Microbiol 1:1–15
Uchida K, Aida KO (1984) An improved method for the glycolate test for simple identification of the acyl type of bacterial cell walls. J Gen Appl Microbiol 30:131–134
Wayne LG, Brenner DJ, Colwell RR, Grimont KAD, Kandler K, other authors, (1987) Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol 37:463–464
Wick RR, Judd LM, Gorrie CL, Holt KE (2017) Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLOS Comput Biol. https://doi.org/10.1371/journal.pcbi.1005595
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67:1613–1617
Acknowledgements
The authors thank Greg Kirby for his assistance with sampling of native plants. We are very grateful to Daniel Jardine for menaquinone analysis and Dr. Lisa O`Donovan of Adelaide Microscopy for SEM visualization.
Funding
This research project is financially supported by Mahasarakham University (Fast Track 2020) Mahasarakham University, Thailand.
Author information
Authors and Affiliations
Contributions
OK and CF planned the experiments. OK carried out the experimental work and prepared the draft manuscript. CF contributed to improve the manuscript and provided the facilities. CS prepared genomic DNA for genome sequencing purposes.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants and/or animals performed by any of the authors. The formal consent is not required in this study.
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The GenBank/EMBL/DDBJ accession number for the 16S rRNA gene sequence of strain CAP 181T is GU434267.
The GenBank/EMBL/DDBJ accession number for the draft genome of strain CAP181T is JAEMUW000000000.
Supplementary Information
Below is the link to the electronic supplementary material.
203_2021_2260_MOESM1_ESM.pptx
Supplementary Figure S1. 16S rRNA gene-based maximum parsimony tree showing the phylogenetic relationships between Micromonospora veneta CAP181T and fourteen strains with valid names belonging to the genus Micromonospora, and Actinoplanes bogorensis NBRC 110975T as the outgroup. Bootstrap values (> 50 %) based on 1000 replicates are shown at the branch nodes and asterisk (*) indicate clades that were conserved in the maximum-likelihood and maximum parsimony trees. (PPTX 844 KB)
203_2021_2260_MOESM2_ESM.pdf
Supplementary Figure S2. Two-dimensional thin-layer chromatography of polar lipids of Micromonospora veneta CAP181T. Chloroform-methanol-water (65:25:4) was used in the first direction, followed by chloroform-acetic acid-methanol-water (40:7.5:6:2) in the second direction. Abbreviations: DPG; Diphosphatidylglycerol, PE; phosphatidylethanolamine, PI; phosphatidylinositol, PIDM; phosphatidylinositol dimannosides, UnGly; unidentified glycolipid, UnA; unidentified lipid with amino group, UnPa; unidentified phospholipid with amino group, B; brown colour, X; starting point. (PDF 544 KB)s
203_2021_2260_MOESM3_ESM.tif
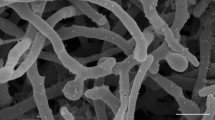
Supplementary Figure S3. Scanning electron microscopy showed hyphae and single spore with hairy surface of strain CAP181T grown on HPDA for 10 days at 27oC. Bar represents 2 μm.s (TIF 342 KB)
Rights and permissions
About this article
Cite this article
Kaewkla, O., Suriyachadkun, C. & Franco, C.M.M. Micromonospora veneta sp. nov., an endophytic actinobacterium with potential for nitrogen fixation and for bioremediation. Arch Microbiol 203, 2853–2861 (2021). https://doi.org/10.1007/s00203-021-02260-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-021-02260-3