Abstract
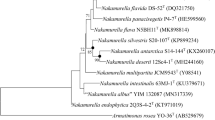
A white-coloured, aerobic, and rod-shaped bacterium, designated strain ID0723T was isolated from evaporator core of automobile air conditioning system. The strain was Gram-stain-negative, catalase positive, oxidase negative, and grew at pH 5.5–9.5, at temperature 18–37 °C, and at 0–2.0% (w/v) NaCl concentration. The phylogenetic analysis and 16S rRNA gene sequence data revealed that the strain ID0723T was affiliated to the genus Schlegelella, with the closest phylogenetic member being Schlegelella brevitalea DSM 7029 T (98.1% sequence similarity). The chemotaxonomic features of strain ID0723T were diphosphatidylglycerol, phosphatidylglycerol, and phosphatidylethanolamine as the main polar lipids; Q-8 as an only ubiquinone; and summed feature 3 (C16:1ω7c and/or C16: 1ω6c), C16:0, and summed feature 8 (C18:1ω7c/or C18:1ω6c) as the major fatty acids. The average nucleotide identity (ANI) and in silico DNA–DNA hybridization values between strain ID0723T and S. brevitalea DSM 7029 T were 74.8% and 20.0%, respectively, which were below the cut-off values of 95% and 70%, respectively. The DNA G + C content was 69.9 mol%. The polyphasic taxonomic data clearly indicated that strain ID0723T represents a novel species in the genus Schlegelella for which the name Schlegelella koreensis sp. nov. is proposed, with the type strain ID0723T (= KCTC 72731 T = NBRC 114611 T).

Similar content being viewed by others
Abbreviations
- NBRC:
-
NITE Biological Resource Center
- KCTC:
-
Korean Collection for Type Cultures
- ANI:
-
Average nucleotide identity
References
Aziz RK, Bartels D, Best AA et al (2008) The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75
Blin K, Shaw S, Steinke K et al (2019) antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res 47:W81–W87
Breznak JA, Costilow RN (2007) Physicochemical factors in growth. In: Reddy CA, Beveridge TJ, Breznak JA et al (eds) Methods for general and molecular bacteriology, 3rd edn. American Society of Microbiology, Washinton, DC, pp 309–329
Chaudhary DK, Kim J (2018) Flavobacterium naphthae sp. nov., isolated from oil-contaminated soil. Int J Syst Evol Microbiol 68:305–309
Chou YJ, Sheu SY, Sheu DS et al (2006) Schlegelella aquatica sp. nov., a novel thermophilic bacterium isolated from a hot spring. Int J Syst Evol Microbiol 56:2793–2797
Collins MD, Jones D (1981) Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implication. Microbiol Rev 45:316–354
Doetsch RN (1981) Determinative methods of light microscopy. In: Gerhardt P, Murray RGE, Costilow RN et al (eds) Manual of methods for general bacteriology. American Society for Microbiology, Washington DC, pp 21–33
Elbanna K, Lütke-Eversloh T, Van Trappen S et al (2003) Schlegelella thermodepolymerans gen. nov., sp. nov., a novel thermophilic bacterium that degrades poly(3-hydroxybutyrate-co-3-mercaptopropionate). Int J Syst Evol Microbiol 53:1165–1168
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution (N Y) 39:783–791
Frank JA, Reich CI, Sharma S et al (2008) Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl Environ Microbiol 74:2461–2470
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Komagata K, Suzuki KI (1988) 4 Lipid and cell-wall analysis in bacterial systematics. Methods Microbiol 19:161–207
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Lee I, Chalita M, Ha S-M et al (2017) ContEst16S: an algorithm that identifies contaminated prokaryotic genomes using 16S RNA gene sequences. Int J Syst Evol Microbiol 67:2053–2057
Lütke-Eversloh T, Elbanna K, Cnockaert MC et al (2004) Caenibacterium thermophilum is later synonym of Schlegelella thermodepolymerans. Int J Syst Evol Microbiol 54:1933–1935
Manaia CM, Nunes OC, Nogales B (2003) Caenibacterium thermophilum gen. nov., sp. nov., isolated from a thermophilic aerobic digester of municipal sludge. Int J Syst Evol Microbiol 53:1375–1382
Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60
Minnikin DE, O’Donnell AG, Goodfellow M et al (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241
Na SI, Kim YO, Yoon SH et al (2018) UBCG: Up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J Microbiol 56:281–285
Pruesse E, Peplies J, Glöckner FO (2012) SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28:1823–1829
Richter M, Rosselló-Móra R (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci 106:19126–19131
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sasser M (1990) Bacterial identification by gas chromatographic analysis of fatty acid methyl esters (GC-FAME). MIDI Tech Note 101 Newwark, MIDI Inc
Smibert RM, Krieg NR (1994) Phenotypic characterization. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR (eds) Methods for general and molecular bacteriology. American Society for Microbiology, Washington, DC, pp 607–654
Tang B, Yu Y, Liang J et al (2019) Reclassification of 'Polyangium brachysporum’ DSM 7029 as schlegelella brevitalea sp. Nov Int J Syst Evol Microbiol 69:2877–2883
Tatusova T, DiCuccio M, Badretdin A et al (2016) NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44:6614–6624
Wayne LG, Brenner DJ, Colwell RR et al (1987) Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Evol Microbiol 37:463–464
Yoon S-H, Ha S, Lim J et al (2017a) A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110:1281–1286
Yoon SH, Ha SM, Kwon S et al (2017b) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67:1613–1617
Zhang Z, Schwartz S, Wagner L, Miller W (2000) A greedy algorithm for aligning DNA sequences. J Comput Biol 7:203–214
Acknowledgements
This work was supported by Rural Development Administration (Project No. PJ014897032020) and Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2019R1A6A3A01091574).
Funding
The GenBank/EMBL/DDBJ Accession number for the 16S rRNA sequence of strain ID0723T is KP326334. The whole genome shotgun sequence of strain ID0723T has been deposited at DDBJ/ENA/GenBank under the Accession JABWMJ000000000. The version described in this paper is version JABWMJ010000000.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest regarding the publication of this manuscript.
Ethical statement
This study does not describe any experimental work related to human.
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chaudhary, D.K., Lee, H., Dahal, R.H. et al. Schlegelella koreensis sp. nov., isolated from evaporator core of automobile air conditioning system. Arch Microbiol 203, 2373–2378 (2021). https://doi.org/10.1007/s00203-021-02206-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-021-02206-9