Abstract
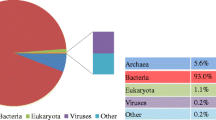
Anaerobic digestion, a recently hot technology to produce biogases especially methane generation for biofuel from wastewater, is considered an effective explanation for energy crisis and global pollution threat. A complex microbiome population is present in sludge, which plays an important role in the digestion of complex polymer into simple monomers. 16S rRNA approaches simply are not enough for amplification due to the involvement of extreme complex population. However, Illumina sequencing is a recent powerful technology to reveal the entire microbiome structure and methane generation pathways in anaerobic digestion. Metagenomic sequencing was tested to reveal the microbial structure of a digested sludge from a local wastewater treatment plant in Beijing. The Illumina HiSeq program was used to extract about 5 GB of data for metagenomic analysis. The classification investigation revealed about 97.64% dominancy of bacteria while 1.78% were detected to be archaea using MG-RAST server. The most abundant bacterial communities were reported to be Actinobacteria, Bacteroidetes, Firmicutes and Proteobacteria. Furthermore, the important microbiome involved in methane generation was revealed. The dominant methanogens were detected (Methanosaeta and Methanosarcina), with affiliation of dominant genes involved in acetoclastic methanogenesis in a digesting sludge. The metagenomic analysis showed that microbial structure and methane generation pathways were successfully dissected in an anaerobic digester.










Similar content being viewed by others
References
Aird D, Ross MG, Chen WS, Danielsson M, Fennell T, Russ C (2011) Analyzing and minimizing PCR amplification bias in Illumina sequencing libraries. Genome Biol 12:18
Albertsen M, Hugenholtz P, Skarshewski A, Nielsen KL, Tyson GW, Nielsen PH (2003) Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol 31:533–538
Albertsen M, Hansen LBS, Saunders AM, Nielsen PH, Nielsen KL (2006) A metagenome of a full-scale microbial community carrying out enhanced biological phosphorus removal. ISME J 6:1094–1106
Amani T, Nosrati M, Sreekrishnan TR (2010) Anaerobic digestion from the viewpoint of microbiological, chemical, and operational aspects—a review. Environ Rev 18:255–278
APHA (1998) Standard methods for the examination of water and wastewater, 20th edn. APHA, Washington
Appels L, Baeyens J, Degreve J, Dewil R (2008) Principles and potential of the anaerobic digestion of waste-activated sludge. Prog Energ Combust 34:755–781
Ariesyady HD, Ito T, Okabe S (2007) Functional bacterial and archaeal community structures of major trophic groups in a full-scale anaerobic sludge digester. Water Res 41:1554–1568
Bragg L, Tyson GW (2014) Metagenomics using next-generation sequencing. In: Paulsen IT, Holmes AJ (eds) Environmental microbiology: methods and protocols. Methods in molecular biology, vol 1096, 2nd edn. Humana Press, New York City, pp 183–201
Canales A, Pareilleux A, Rols JL, Goma G, Huyard A (1994) Decreased sludge production strategy for domestic wastewater treatment. Water Sci Technol 30:97–106
Ferry JG (1999) Enzymology of one-carbon metabolism in methanogenic pathways. FEMS Microbiol Rev 23:13–38
Garcia-Peña EI, Parameswaran P, Kang DW, Canul-Chan M, Krajmalnik-Brown R (2011) Anaerobic digestion and co-digestion processes of vegetable and fruit residues: process and microbial ecology. Bioresour Technol 102:9447–9455
Glenn TC (2011) Field guide to next-generation DNA sequencers. Mol Ecol Resour 11:759–769
Guo J, Peng Y, Wang S, Ma B, Ge S, Wang Z (2013) Pathways and organisms involved in ammonia oxidation and nitrous oxide emission. Crit Rev Environ Sci Technol 43:2213–2296
Huson D, Mitra S, Ruscheweyh H, Weber N, Schuster S (2011) Integrative analysis of environmental sequences using MEGAN4. Genome Res 21:1552–1560
Ju F, Guo F, Ye L, Xia Y, Zhang T (2014) Metagenomic analysis on seasonal microbial variations of activated sludge from a full-scale wastewater treatment plant over 4 years. Environ Microbiol Rep 6:80–89
Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S (2006) From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res 34:354–357
Li A, Chu Y, Wang X, Ren L, Yu J, Liu X (2013) A pyrosequencing-based metagenomic study of methane-producing microbial community in solid-state biogas reactor. Biotechnol Biofuels 6:3
Liu Y, Whitman WB, Wiegel J, Maier RJ, Adams MWW (eds) (2008) Incredible anaerobes: from physiology to genomics to fuels, vol 1125. HighWire Press, New York, pp 171–189
Mackelprang R, Waldrop MP, DeAngelis KM, David MM, Chavarria KL, Blazewicz SJ (2011) Metagenomic analysis of a permafrost microbial community reveals a rapid response to thaw. Nature 480:368-U120
Mardis ER (2008) The impact of next-generation sequencing technology on genetics. Trends Genet 24:133–141
Mason OU, Scott NM, Gonzalez A, Robbins-Pianka A, Baelum J, Kimbrel J (2014) Metagenomics reveals sediment microbial community response to Deepwater Horizon oil spill. ISME 8:1464–1475
Meyer F, Paarmann D, D’Souza M, Olson R, Glass EM, Kubal M (2008) The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9:386
Mitra S, Rupek P, Richter DC, Urich T, Gilbert JA, Meyer F (2011) Functional analysis of metagenomes and metatranscriptomes using SEED and KEGG. BMC Bioinformatics 12:21
Nelson MC, Morrison M, Yu Z (2011) A meta-analysis of the microbial diversity observed in anaerobic digesters. Bioresour Technol 102:3730–3739
Noor E, Eden E, Milo R, Alon U (2010) Central carbon metabolism as a minimal biochemical walk between precursors for biomass and energy. Mol Cell 39:809–820
Papagianni M (2012) Recent advances in engineering the central carbon metabolism of industrially important bacteria. Microb Cell Fact 11:50
Pelletier E, Kreimeyer A, Bocs S, Rouy Z, Gyapay G, Chouari R (2008) “Candidatus Cloacamonas acidaminovorans”: genome sequence reconstruction provides a first glimpse of a new bacterial division. J Bacteriol 190:2572–2579
Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C et al (2010) A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59-U70
Ragsdale SW, Pierce E (2008) Acetogenesis and the Wood–Ljungdahl pathway of CO2 fixation. Biochim Biophys Acta Proteins Proteom 1784:1873–1898
Ramsay IR, Pullammanappallil PC (2001) Protein degradation during anaerobic wastewater treatment: derivation of stoichiometry. Biodegradation 12:247–257
Regueiro L, Veiga P, Figueroa M, Alonso-Gutierrez J, Stams AJM, Lema JM (2012) Relationship between microbial activity and microbial community structure in six full-scale anaerobic digesters. Microbiol Res 167:581–589
Sandoval LCJ, Vergara MM, De Carreno AM, Castillo MEF (2009) Microbiological characterization and specific methanogenic activity of anaerobe sludges used in urban solid waste treatment. Waste Manag 29:704–711
Sundberg C, Al-Soud WA, Larsson M, Alm E, Yekta SS, Svensson BH (2013) 454 pyrosequencing analyses of bacterial and archaeal richness in 21 full-scale biogas digesters. FEMS Microbiol Ecol 85:612–626
Tatusov RL, Galperin MY, Natale DA, Koonin EV (2000) The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res 28:33–36
Traversi D, Villa S, Lorenzi E, Degan R, Gilli G (2012) Application of a real-time qPCR method to measure the methanogen concentration during anaerobic digestion as an indicator of biogas production capacity. J Environ Manag 111:173–177
van Loosdrecht MCM, Brdjanovic D (2014) Anticipating the next century of wastewater treatment. Science 344:1452–1453
Vanwonterghem I, Jensen PD, Ho DP, Batstone DJ, Tyson GW (2014a) Linking microbial community structure, interactions and function in anaerobic digesters using new molecular techniques. Curr Opin Biotechnol 27:55–64
Vanwonterghem I, Jensen PD, Dennis PG, Hugenholtz P, Rabaey K, Tyson GW (2014b) Deterministic processes guide long-term synchronised population dynamics in replicate anaerobic digesters. ISME 8:2015–2028
Wong MT, Zhang D, Li J, Hui RKH, Tun HM, Brar MS (2013) Towards a metagenomic understanding on enhanced biomethane production from waste activated sludge after pH 10 pretreatment. Biotechnol Biofuels 6:38
Yang Y, Yu K, Xia Y, Lau FTK, Tang DTW, Fung WC (2014) Metagenomic analysis of sludge from full-scale anaerobic digesters operated in municipal wastewater treatment plants. Appl Microbiol Biot 98:5709–5718
Ye L, Zhang T, Wang TT, Fang ZW (2012) Microbial structures, functions, and metabolic pathways in wastewater treatment bioreactors revealed using high-throughput sequencing. Environ Sci Technol 46:13244–13252
Yu Y, Lee C, Hwang S (2005) Analysis of community structures in anaerobic processes using a quantitative real-time PCR method. Water Sci Technol 52:85–91
Ziganshin AM, Liebetrau J, Proeter J, Kleinsteuber S (2013) Microbial community structure and dynamics during anaerobic digestion of various agricultural waste materials. Appl Microbiol Biotechnol 97:5161–5174
Acknowledgements
This work was supported by Major Science and Technology Program for Water Pollution Control and Treatment of China (Grant no. 2017ZX07102-004), and National Natural Science Foundation of China (Grant no. 21206084). We are grateful to School of Medicine, Tsinghua University for providing the tools of metagenomic Matlab.
Author information
Authors and Affiliations
Contributions
All the authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declared that there is no conflict of interest.
Additional information
Communicated by Kristina Beblo-Vranesevic.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ali, N., Gong, H., Liu, X. et al. Evaluation of bacterial association in methane generation pathways of an anaerobic digesting sludge via metagenomic sequencing. Arch Microbiol 202, 31–41 (2020). https://doi.org/10.1007/s00203-019-01716-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-019-01716-x