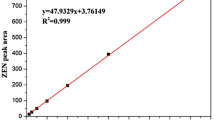
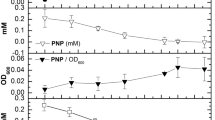
Abstract
Bisphenol A (BPA) is a synthetic chemical with known deleterious effects on biota. A genome sequencing project is an important starting point for designing a suitable BPA bioremediation process, because it provides valuable genomic information about the physiological, metabolic, and genetic potential of the microbes used for the treatment. This study explored genomic insights provided by the BPA-degrading strain Bacillus sp. GZB, previously isolated from electronic-waste-dismantling site. The GZB genome is a circular chromosome, comprised of a total of 4,077,007 bp with G+C content comprising 46.2%. Genome contained 23 contigs encoded by 3881 protein-coding genes with nine rRNA and 53 tRNA genes. A comparative study demonstrated that strain GZB bloomed with some potential features as compared to other Bacillus species. In addition, strain GZB developed spore cells and displayed laccase activity while growing at elevated stress levels. Most importantly, strain GZB contained many protein-coding genes associated with BPA degradation, as well as the degradation of several other compounds. The protein-coding genes in the genome revealed the genetic mechanisms associated with the BPA degradation by strain GZB. This study predicts four possible degradation pathways for BPA, contributing to the possible use of strain GZB to remediate different polluted environments in the future.





Similar content being viewed by others
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Angiuoli SV, Gussman A, Klimke W, Cochrane G, Field D, Garrity G, Kodira CD, Kyrpides N, Madupu R, Markowitz V, Tatusova T, Thomson N, White O (2008) Toward an online repository of standard operating procedures (SOPs) for (Meta) genomic annotation. Omics J Integrative Bio 12:137–141
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppiq JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G (2000) Gene ontology: tool for the unification of biology. Nat Genet 25:25–29
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil KL, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O (2008) The RAST server: rapid annotations using subsystems technology. BMC Genom 9:75–89
Bertelli C, Laird MR, Williams KP, Fraser S, Lau BY, Hoad G, Winsor GL, Brinkman FSL (2017) IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res 45:W30–W35
Binnewies TT, Motro Y, Hallin PF, Lund O, Dunn D, La T, Hampson DJ, Bellgard M, Wassenaar TM, Ussery DW (2006) Ten years of bacterial genome sequencing: comparative-genomics-based discoveries. Funct Integr Genomics 6:165–185
Bressuire-Isoard C, Bornard I, Henriques AO, Carlin F, Broussolle V (2016) Sporulation temperature reveals a requirement for CotE in the assembly of both the coat and exosporium layers of Bacillus cereus spores. Appl Environ Microbiol 82:232–243
Chen XH, Koumoutsi A, Scholz R, Eisenreich A, Schneider K, Heinemeyer I, Morgenstern B, Voss B, Hess WR, Reva O, Junge H, Voigt B, Jungblut PR, Vater J, Süssmuth R, Liesegang H, Strittmatter A, Gottschalk G, Borriss R (2007) Comparative analysis of the complete genome sequence of the plant growth-promoting bacterium Bacillus amyloliquefaciens FZB42. Nat Biotechnol 25:1007
Chen D, Kannan K, Tan H, Zheng Z, Feng YL, Wu Y, Widelka M (2016) Bisphenol analogs other than BPA: environmental occurrence, human exposure, and toxicity—a review. Environ Sci Technol 50:5438–5453
Crow A, Lewin A, Hecht O, Carlsson Moller M, Moore GR, Hederstedt L, Le Brun NE (2009) Crystal structure and biophysical properties of Bacillus subtilis BdbD. An oxidizing thiol:disulfide oxidoreductase containing a novel metal site. J Biol Chem 284:23719–23733
Das R, Kazy SK (2014) Microbial diversity, community composition and metabolic potential in hydrocarbon contaminated oily sludge: prospects for in situ bioremediation. Environ Sci Pollut Res 21:7369–7389
Das R, Li G, Mai B, An T (2018)) Spore cells from BPA degrading bacteria Bacillus sp. GZB displaying high laccase activity and stability for BPA degradation. Sci Total Environ 640–641:798–806
Degtyarenko KN (1995) Structural domains of P450-containing monooxygenase systems. Protein Eng 8:737–747
Dwivedi UN, Singh P, Pandey VP, Kumar A (2011) Structure–function relationship among bacterial, fungal and plant laccases. J Mol Catal B: Enzyme 68:117–128
Enguita FJ, Martins LO, Henriques AO, Carrondo MA (2003) Crystal structure of a bacterial endospore coat component. A laccase with enhanced thermostability properties. J Biol Chem 278:19416–19425
Feng X, Coulombe PA (2015) A role for disulfide bonding in keratin intermediate filament organization and dynamics in skin keratinocytes. J Cell Biol 209:59–72
Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, Ashburner M, Axelrod N, Baldauf S, Ballard S, Boore J, Cochrane G, Cole J, Dawyndt P, De Vos P, dePamphilis C, Edwards R, Faruque N, Feldman R, Gilbert J, Gilna P, Glöckner FO, Goldstein P, Guralnick R, Haft D, Hancock D, Hermjakob H, Hertz-Fowler C, Hugenholtz P, Joint I, Kagan L, Kane M, Kennedy J, Kowalchuk G, Kottmann R, Kolker E, Kravitz S, Kyrpides N, Leebens-Mack J, Lewis SE, Li K, Lister AL, Lord P, Maltsev N, Markowitz V, Martiny J, Methe B, Mizrachi I, Moxon R, Nelson K, Parkhill J, Proctor L, White O, Sansone SA, Spiers A, Stevens R, Swift P, Taylor C, Tateno Y, Tett A, Turner S, Ussery D, Vaughan B, Ward N, Whetzel T, Gil IS, Wilson G, Wipat A (2008) The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol 26:541–547
Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, Salazar GA, Tate J, Bateman A (2015) The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44:D279–D285
García-Díaz C, Ponce-Noyola MT, Esparza-García F, Rivera-Orduña F, Barrera-Cortés J (2013) PAH removal of high molecular weight by characterized bacterial strains from different organic sources. Int Biodeter Biodegr 85:311–322
Gerischer U (2002) Specific and global regulation of genes associated with the degradation of aromatic compounds in bacteria. J Mol Microbiol Biotechnol 4:111–121
Giebel HA, Klotz F, Voget S, Poehlein A, Grosser K, Teske A, Brinkhoff T (2016) Draft genome sequence of the marine Rhodobacteraceae strain O3.65, cultivated from oil-polluted seawater of the Deepwater Horizon oil spill. Stand Genomic Sci 11:81
Glockler R, Tschech A, Fuchs G (1989) Reductive dehydroxylation of 4-hydroxybenzoyl-CoA to benzoyl-CoA in a denitrifying, phenol-degrading Pseudomonas species. FEBS Lett 251:237–240
Grignard E, Lapenna S, Bremer S (2012) Weak estrogenic transcriptional activities of Bisphenol A and Bisphenol S. Toxicol In Vitro 26:727–731
Hacker J, Kaper JB (2000) Pathogenicity islands and the evolution of microbes. Ann Rev Microbiol 54:641–679
Hautphenne C, Penninckx M, Debaste F (2016) Product formation from phenolic compounds removal by laccases: a review. Environ Technol Innovation 5:250–266
Held C, Kandelbauer A, Schroeder M, Cavaco-Paulo A, Gübitz GM (2005) Biotransformation of phenolics with laccase containing bacterial spores. Environ Chem Lett 3:74–77
Hong YH, Ye CC, Zhou QZ, Wu XY, Yuan JP, Peng J, Deng H, Wang JH (2017) Genome sequencing reveals the potential of Achromobacter sp. HZ01 for bioremediation. Front Microbiol 7:1507
Huang YQ, Wong CKC, Zheng JS, Bouwman H, Barra R, Wahlström B, Neretin L, Wong MH (2012) Bisphenol A (BPA) in China: a review of sources, environmental levels, and potential human health impacts. Environ Int 42:91–99
Im J, Loffler FE (2016) Fate of bisphenol A in terrestrial and aquatic environments. Environ Sci Technol 50:8403–8416
Imai M, Shimada H, Watanabe Y, Matsushima-Hibiya Y, Makino R, Koga H, Horiuchi T, Ishimura Y (1989) Uncoupling of the cytochrome P-450cam monooxygenase reaction by a single mutation, threonine-252 to alanine or valine: possible role of the hydroxy amino acid in oxygen activation. Proc Natl Acad Sci USA 86:7823–7827
Jouanneau Y, Meyer C, Duraffourg N (2016) Dihydroxylation of four-and five-ring aromatic hydrocarbons by the naphthalene dioxygenase from Sphingomonas CHY-1. Appl Microbiol Biotechnol 100:1253–1263
Jukes TH, Cantor CR (1969) Evolution of protein molecules. In: Munro HN (ed) Mammalian protein metabolism. Academic Press., New York, pp 21–132
Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M (2016) KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 44:D457–D462
Kim SJ, Kweon O, Jones RC, Freeman JP, Edmondson RD, Cerniglia CE (2007) Complete and integrated pyrene degradation pathway in Mycobacterium vanbaalenii PYR-1 based on systems biology. J Bacteriol 189:464–472
Kolvenbach B, Schlaich N, Raoui Z, Prell J, Zuhlke S, Schaffer A, Guengerich FP, Corvini PFX (2007) Degradation pathway of bisphenol A: Does ipso substitution apply to phenols containing a quaternary α-carbon structure in the para position? Appl Environ Microbiol 73:4776–4784
Krogh A, Larsson B, Von Heijne G, Sonnhammer EL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580
Lagesen K, Hallin P, Rødland EA, Stærfeldt HH, Rognes T, Ussery DW (2007) RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 35:3100–3108
Larsson K, Lindh CH, Jönsson BAG, Giovanoulis G, Bibi M, Bottai M, Bergström A, Berglund M (2017) Phthalates, non-phthalate plasticizers, and bisphenols in Swedish preschool dust in relation to children’s exposure. Environ Int 102:114–124
Li G, Zu L, Wong PK, Hui X, Lu Y, Xiong J, An T (2012) Biodegradation and detoxification of Bisphenol A with one newly-isolated strain Bacillus sp. GZB: kinetics, mechanism, and estrogenic transition. Bioresour Technol 114:224–230
Li ZY, Wu YH, Huo YY, Cheng H, Wang CS, Xu XW (2016) Complete genome sequence of a benzo[a]pyrene-degrading bacterium Altererythrobacter epoxidivorans CGMCC 1.7731T. Mar Genomics 25:39–41
Li C, Lu Q, Ye J, Qin H, Long Y, Wang L, Ou H (2018) Metabolic and proteomic mechanism of bisphenol A degradation by Bacillus thuringiensis. Sci Total Environ 640–641:714–725
Liang WX, Deutscher MP (2013) Ribosomes regulate the stability and action of the exoribonuclease RNase R. J Biol Chem 288:34791–34798
Liang Z, Li G, An T, Das R (2016) Draft genome sequence of Bacillus sp. GZT, a 2,4,6-tribromophenol degrading strain isolated from the river sludge of an electronic waste-dismantling region. Genome Announc 3:e00474–e00416
Liang Z, Li G, An T (2017) Purifying, cloning and characterizing a novel dehalogenase from Bacillus sp. GZT to enhance the biodegradation of 2,4,6-tribromophenol in water. Environ Pollut 225:104–111
Little S, Driks A (2001) Functional analysis of the Bacillus subtilis morphogenetic spore coat protein CotE. Mol Microbiol 42:1107–1120
Logan NA, Vos PD (2015) Bacillus. In: Whitman WB (ed) Bergey’s manual of systematics of archaea and bacteria. Wiley, Hoboken
Lu L, Zhao M, Wang TN, Zhao LY, Du MH, Li TL, Li DB (2012) Characterization and dye decolorization ability of an alkaline resistant and organic solvents tolerant laccase from Bacillus licheniformis LS04. Bioresour Technol 115:35–40
Lu A, Peng Q, Ling E (2014) Formation of disulfide bonds in insect prophenoloxidase enhances immunity through improving enzyme activity and stability. Dev Comp Immunol 44:351–358
Luo A, Wu YR, Xu Y, Kan J, Qiao J, Liang L, Huang T, Hu Z (2016) Characterization of a cytochrome P450 monooxygenase capable of high molecular weight PAHs oxidization from Rhodococcus sp. P14. Process Biochem 51:2127–2133
Madigan MT, Martinko J (1997) Brock biology of microorganisms. Pearson Prentice Hall Inc., In
McKenney PT, Driks A, Eichenberger P (2013) The Bacillus subtilis endospore: assembly and functions of the multilayered coat. Nat Rev Microbiol 11:33–44
McLeod MP, Warren RL, Hsiao WW, Araki N, Myhre M, Fernandes C, Miyazawa D, Wong W, Lillquist AL, Wang D, Dosanjh M, Hara H, Petrescu A, Morin RD, Yang G, Stott JM, Schein JE, Shin H, Smailus D, Siddiqui AS, Marra MA, Jones SJ, Holt R, Brinkman FS, Miyauchi K, Fukuda M, Davies JE, Mohn WW, Eltis LD (2006) The complete genome of Rhodococcus sp. RHA1 provides insights into a catabolic powerhouse. Proc Natl Acad Sci USA 103:15582–15587
Meier-Kolthoff JP, Klenk HP, Göker M (2014) Taxonomic use of DNA G + C content and DNA-DNA hybridization in the genomic age. Int J Syst Evol Microbiol 64:352–356
Mouttaki H, Johannes J, Meckenstock RU (2012) Identification of naphthalene carboxylase as a prototype for the anaerobic activation of non-substituted aromatic hydrocarbons. Environ Microbiol 14:2770–2774
Nicholson WL, Munakata N, Horneck G, Melosh HJ, Setlow P (2000) Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol Mol Biol Rev 64:548–572
Pal S, Kundu A, Das Banerjee T, Mohapatra B, Roy A, Manna R, Sar P, Kazy SK (2017) Genome analysis of crude oil degrading Franconibacter pulveris strain DJ34 revealed its genetic basis for hydrocarbon degradation and survival in oil contaminated environment. Genomics 109:374–382
Peng R, Shi B, Fu X, Tian Y, Zhao W, Zhu B, Xu J, Han H, Yuan Z, Liu K, Yao Q (2015) Improving rice ability to degrade different polycyclic aromatic hydrocarbons through multigene transformation of a hybrid dioxygenase system. Plant Mol Biol Report 33:1030–1041
Petersen TN, Brunak S, von Heijne G, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Med 8:785–786
Prokka ST (2014) Rrapid prokaryotic genome annotation. Bioinformatics 30:2068–2069
Rochester JR, Bolden AL (2015) Bisphenol S and F: a systematic review and comparison of the hormonal activity of Bisphenol A substitutes. Environ Health Perspect 123:643–650
Rodriguez -RLM, Konstantinidis KT (2014) Bypassing cultivation to identify bacterial species. Microbe 9:111–118
Rojo F (2009) Degradation of alkanes by bacteria: mini review. Environ Microbiol 11:2477–2490
Sasaki M, Akahira A, Tsuchido T, Matsumura Y (2005) Purification of cytochrome P450 and ferredoxin involved in BPA degradation by Sphingomonas sp. strain AO1. Appl Environ Microbiol 171:8024–8030
Sasaki M, Tsuchido T, Matsumura Y (2008) Molecular cloning and characterization of cytochrome P450 and ferredoxin genes involved in bisphenol A degradation in Sphingomonas bisphenolicum strain AO1. Appl Microbiol 105:1158–1169
Sophos NA, Vasiliou V (2003) Aldehyde dehydrogenase gene superfamily: the 2002 update. Chem Biol Interact 143–144:5–22
Spivack J, Leib TK, Lobos JH (1994) Novel pathway for bacterial metabolism of bisphenol A. Rearrangements and stilbene cleavage in bisphenol A metabolism. J Biol Chem 269:7323–7329
Sudtachat N, Ito N, Itakura M, Masuda S, Eda S, Mitsui H, Kawaharada Y, Minamisawa K (2009) Aerobic vanillate degradation and C1 compound metabolism in Bradyrhizobium japonicum. Appl Environ Microbiol 75:5012–5017
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729
Telke AA, Kalyani DC, Jadhav UU, Parshetti GK, Govindwar SP (2009) Purification and characterization of an extracellular laccase from a Pseudomonas sp. LBC1 and its application for the removal of bisphenol A. J Mol Catal B-Enzym 61:252–260
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucleic Acids Res 22:4673–4680
Wang Y, Xin Y, Cao X, Xue S (2015) Enhancement of L-2-haloacid dehalogenase expression in Pseudomonas stutzeri DEH138 based on the different substrate specificity between dehalogenase-producing bacteria and their dehalogenases. World J Microbiol Biotechnol 31:669–673
Wu M, Pan C, Chen Z, Jiang L, Lei P, Yang M (2017) Bioconcentration pattern and induced apoptosis of Bisphenol A in zebrafish embryos at environmentally relevant concentrations. Environ Sci Pollut R 24:6611–6621
Yang R, Liu G, Chen T, Li S, An L, Zhang G, Li G, Chang S, Zhang W, Chen X, Wu X, Zhang B (2018) Characterization of the genome of a Nocardia strain isolated from soils in the Qinghai–Tibetan Plateau that specifically degrades crude oil and of this biodegradation. Genomics. https://doi.org/10.1016/j.ygeno.2018.02.010
Yoon SH, Ha SM, Lim JM, Kwon SJ, Chun J (2017) A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110:1281–1286
Zhang C, Zhang S, Diao H, Zhao H, Zhu X, Lu F, Lu Z (2013) Purification and characterization of a temperature-and pH-stable laccase from the spores of Bacillus vallismortis fmb-103 and its application in the degradation of malachite green. J Agric Food Chem 61:5468–5473
Acknowledgements
We gratefully acknowledge Sangon Biotech, Shanghai, China for genome analysis. This study was financially supported by the National Natural Science Foundation of China (41877363, 41425015 and 41373103) and the Science and Technology Program of Guangzhou, China (201704020185).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of affairs concerning the work published in this paper.
Additional information
Communicated by Erko Stackebrandt.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Das, R., Liang, Z., Li, G. et al. Genome sequence of a spore-laccase forming, BPA-degrading Bacillus sp. GZB isolated from an electronic-waste recycling site reveals insights into BPA degradation pathways. Arch Microbiol 201, 623–638 (2019). https://doi.org/10.1007/s00203-019-01622-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-019-01622-2