Abstract
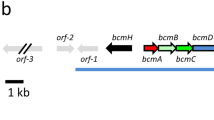
Carbomycins are 16-membered macrolide antibiotics produced by Streptomyces thermotolerans ATCC 11416T. To characterize gene cluster responsible for carbomycin biosynthesis, the draft genome sequences for strain ATCC 11416T were obtained, from which the partial carbomycin biosynthetic gene cluster was identified. This gene cluster was approximately 40 kb in length, and encoding 30 ORFs. Two putative transcriptional regulatory genes, acyB2 and cbmR, were inactivated by insertion of the apramycin resistance gene, and the resulting mutants were unable to produce carbomycin, thus confirming the involvement of two regulatory genes in carbomycin biosynthesis. Overexpression of acyB2 greatly improved the yield of carbomycin; however, overexpression of cbmR blocked carbomycin production. The qPCR analysis of the carbomycin biosynthetic genes in various mutants indicated that most genes were highly expressed in acyB2-overexpressing strains, but few expressed in cbmR-overexpressing strains. Furthermore, acyB2 co-expression with 4″-isovaleryltransferase gene (ist), resulted in efficient biotransformation of spiramycin into bitespiramycin in S. lividans TK24, whereas ist gene regulated by acyB2 and cbmR would cause the lower efficiency of spiramycin biotransformation. These results indicated that AcyB2 was a pathway-specific positive regulator of carbomycin biosynthesis. However, CbmR played a dual role in the carbomycin biosynthesis by acting as a positive regulator, and as a repressor at cbmR high expression levels.




Similar content being viewed by others
References
Arisawa A, Kawamura N, Tsunekawa H, Okamura K, Tone H, Okamoto R (1993) Cloning and nucleotide sequences of two genes involved in the 4″-O-acylation of macrolide antibiotics from Streptomyces thermotolerans. Biosci Biotechnol Biochem 57:2020–2025
Arisawa A, Kawamura N, Takeda K, Tsunekawa H, Okamura K, Okamoto R (1994) Cloning of the macrolide antibiotic biosynthesis gene acyA, which encodes 3-O-acyltransferase, from Streptomyces thermotolerans and its use for direct fermentative production of a hybrid macrolid e antibiotic. Appl Environ Microbiol 60:2657–2660
Arisawa A, Tsunekawa H, Okamura K, Okamoto R (1995) Nucleotide sequence analysis of the carbomycin biosynthetic genes including the 3-O-acyltransferase gene from Streptomyces thermotolerans. Biosci Biotechnol Biochem 59:582–588
Arisawa A, Kawamura N, Narita T, Kojima I, Okamura K, Tsunekawa H, Yoshioka T, Okamoto R (1996) Direct fermentative production of acyltylosins by genetically-engineered strains of Streptomyces fradiae. J Antibiot 49:349–354
Bush MJ, Tschowri N, Schlimpert S, Flärdh K, Buttner MJ (2015) c-di-GMP signalling and the regulation of developmental transitions in streptomycetes. Nat Rev Microbiol 13:749–760
Cundliffe E, Bate N, Butler A, Fish S, Gandecha A, Merson Davies L (2001) The tylosin-biosynthetic genes of Streptomyces fradiae. Antonie Van Leeuwenhoek 79:229–234
Epp JK, Burgett SG, Schoner BE (1987) Cloning and nucleotide sequence of a carbomycin-resistance gene from Streptomyces thermotolerans. Gene 53:73–83
Epp JK, Huber ML, Turner JR, Goodson T, Schoner BE (1989) Production of a hybrid macrolide antibiotic in Streptomyces ambofaciens and Streptomyces lividans by introduction of a cloned carbomycin biosynthetic gene from Streptomyces thermotolerans. Gene 85:293–301
Hansen JL, Ippolito JA, Ban N, Nissen P, Moore PB, Steitz TA (2002) The structures of four macrolide antibiotics bound to the large ribosomal subunit. Mol Cell 10:117–128
Karray F, Darbon E, Oestreicher N, Dominquez H, Tuphile K, Gagnat J, Blondelet-Rouault MH, Gerbaud C, Pernodet JL (2007) Organization of the biosynthetic gene cluster for the macrolide antibiotic spiramycin in Streptomyces ambofaciens. Microbiology 153:4111–4122
Karray F, Darbon E, Nguyen HC, Gagnat J, Pernodet JL (2010) Regulation of the biosynthesis of the macrolide antibiotic spiramycin in Streptomyces ambofaciens. J Bacteriol 192:5813–5821
Kiyoshima K, Sakamoto M, Nomura H, Yoshioka T, Okamoto R, Sawa T, Naganawa H, Takeuchi T (1989) Structure-activity relationship studies on 4″-O-acyltylosin derivatives: significance of their 23-O-mycinosyl and 4″-O-acyl moieties in antimicrobial activity against macrolide-resistant microbes. J Antibiot 42:1661–1672
Menninger JR, Otto DP (1982) Erythromycin, carbomycin, and spiramycin inhibit protein synthesis by stimulating the dissociation of peptidyl-tRNA from ribosomes. Antimicrob Agents Chemother 21:811–818
Mo HB, Bai LQ, Wang SL, Yang KQ (2004) Construction of efficient conjugal plasmids between Escherichia coli and streptomycetes. Sheng Wu Gong Cheng Xue Bao 20(5):662–666
Nguyen HC, Karray F, Lautru S, Gagnat J, Lebrihi A, Huynh TD, Pernodet JL (2010) Glycosylation steps during spiramycin biosynthesis in Streptomyces ambofaciens: involvement of three glycosyltransferases and their interplay with two auxiliary proteins. Antimicrob Agents Chemother 54:2830–2839
Okamoto R, Fukumoto T, Nomura H, Kiyoshima K, Nakamura K, Takamatsu A, Naganawa H, Takeuchi T, Umezawa H (1980a) Physico-chemical properties of new acyl derivatives of tylosin produced by microbial transformation. J Antibiot 33:1300–1308
Okamoto R, Tsuchiya M, Nomura H, Iguchi H, Kiyoshima K, Hori S, Inui T, Sawa T, Takeuchi T, Umezawa H (1980b) Biological properties of new acyl derivatives of tylosin. J Antibiot 33:1309–1315
Omura S, Sano H, Sunazuka T (1985) Structure activity relationships of spiramycins. J Antimicrob Chemother 16:1–11
Romero-Rodríguez A, Robledo-Casados I, Sánchez S (2015) An overview on transcriptional regulators in Streptomyces. Biochim Biophys Acta 1849:1017–1039
Sambrook J, Russell DW (2001) In molecular cloning: a laboratory manual, 3rd edn. Cold Spring, Cold Spring Harbor
Sano H, Sunazuka T, Tanaka H, Yamashita K, Okachi R, Omura S (1984) Chemical modification of spiramycins. IV. Synthesis and in vitro and in vivo activities of 3″,4″-diacylated and 3,3″,4″-triacylates of spiramycin I. J Antibiot 37:760–772
Shang GD, Dai JL, Wang YG (2001) Construction and physiological studies on a stable bioengineered strain of shengjimycin. J Antibiot 54:66–73
Shi XG, Sun YM, Zhang YF, Zhong DF (2004) Tissue distribution of bitespiramycin and spiramycin in rats. Acta Pharmaco Sin 25:1396–1401
Tobias K, Bibb MJ, Mark JB (2000) Practical streptomyces genetics. The John Innes Foundation, Norwich
Tschowri N, Schumacher MA, Schlimpert S, Chinnam NB, Findlay KC, Brennan RG, Buttner MJ (2014) Tetrameric c-di-GMP mediates effective transcription factor dimerization to control Streptomyces development. Cell 158:1136–1147
Vara J, Lewandowska-Skarbek M, Wang YG, Donadio S, Hutchinson CR (1989) Cloning of genes governing the deoxysugar portion of the erythromycin biosynthesis pathway in Saccharopolyspora erythraea (Streptomyces erythreus). J Bacteriol 171:5872–5881
Wagner RL, Hochstein FA, Murai K, Messina N, Regna PP (1953) Magnamycin, a new antibiotic. J Am Chem Soc 75:4684–4687
Zhang JH, Zhong JJ, Dai JL, Wang Y, Xia H, He W (2014) Expression of 4″-O-isovaleryltransferase gene from Streptomyces thermotolerans in Streptomyces lividans TK24. Sheng Wu Gong Cheng Xue Bao 30:1390–1400
Zhong D, Shi X, Sun L, Chen X (2003) Determination of three major components of bitespiramycin and their major active metabolites in rat plasma by liquid chromatography-ion trap mass spectrometry. J Chromatogr B Anal Technol Biomed Life Sci 791(1–2):45–53
Acknowledgements
This work was supported in part by the National Natural Science Foundation Grant 81172972 to W.-Q.H. and by the National Science and Technology Major Project 2014ZX09201003-002 from the Ministry of Science and Technology to W.-Q.H.
Author information
Authors and Affiliations
Contributions
WH conceived and designed the experiments. JZ, ZL, JD, WH performed the experiments. WH analyzed the data, contributed reagents/materials/analysis tools and wrote the paper.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Additional information
Communicated by Erko Stackebrandt.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhong, J., Lu, Z., Dai, J. et al. Identification of two regulatory genes involved in carbomycin biosynthesis in Streptomyces thermotolerans . Arch Microbiol 199, 1023–1033 (2017). https://doi.org/10.1007/s00203-017-1376-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-017-1376-z