Abstract
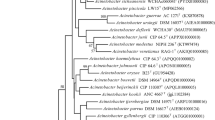
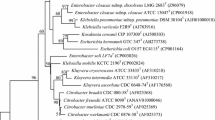
Strain THG–SQM11T, a Gram-negative, aerobic, non-motile, coccus-shaped bacterium, was isolated from wheat seedlings plant in P. R. China. Strain THG–SQM11T was closely related to members of the genus Acinetobacter and showed the highest 16S rRNA sequence similarities with Acinetobacter junii (97.9 %) and Acinetobacter kookii (96.1 %). DNA–DNA hybridization showed 41.3 ± 2.4 % DNA reassociation with A. junii KCTC 12416T. Chemotaxonomic data revealed that strain THG–SQM11T possesses ubiquinone-9 as the predominant respiratory quinone, C18:1 ω9c, summed feature 3 (C16:1 ω7c and/or C16:1 ω6c), and C16:0 as the major fatty acids. The major polar lipids were found to be diphosphatidylglycerol, phosphatidylethanolamine, phosphatidylglycerol, and phosphatidylcholine. The DNA G+C content was 41.7 mol %. These data, together with phenotypic characterization, suggest that the isolate represents a novel species, for which the name Acinetobacter plantarum sp. nov. is proposed, with THG–SQM11T as the type strain (=CCTCC AB 2015123T =KCTC 42611T).

Similar content being viewed by others
References
Anandham R, Weon HY, Kim SJ, Kim YS, Kim BY, Kwon SW (2010) Acinetobacter brisouii sp. nov., isolated from a wetland in Korea. J Microbiol 48:36–39
Carr EL, Kämpfer P, Patel BKC, Gürtler V, Seviour RJ (2003) Seven novel species of Acinetobacter isolated from activated sludge. Int J Syst Evol Microbiol 53:953–963
Choi JY, Ko G, Jheong W, Huys G, Seifert H, Dijkshoorn L, Ko KS (2013) Acinetobacter kookii sp. nov., isolated from soil. Int J Syst Bacteriol 63(12):4402–4406
Collins MD, Jones D (1981) Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implications. Microbiol Rev 45:316–354
Dijkshoorn L, Nemec A, Seifert H (2007) An increasing threat in hospitals: multidrug–resistant Acinetobacter baumannii. Nat Rev Microbiol 5:939–951
Ezaki T, Hashimoto Y, Yabuuchi E (1989) Fluorometric deoxyribonucleic acid–deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int J Syst Bacteriol 39:224–229
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Felsenstein J (1985) Confidence limit on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Feng GD, Yang SZ, Wang YH, Deng MR, Zhu HH (2014) Acinetobacter guangdongensis sp. nov., isolated from abandoned lead–zinc ore. Int J Syst Bacteriol 64:3417–3421
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Syst Zool 20:406–416
Hall TA (1999) BioEdit: a user–friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hiraishi A, Ueda Y, Ishihara J, Mori T (1996) Comparative lipoquinone analysis of influent sewage and activated sludge by high–performance liquid chromatography and photodiode array detection. J Gen Appl Microbiol 42:457–469
Juni E (1972) Interspecies transformation of Acinetobacter: genetic evidence for a ubiquitous genus. J Bacteriol 112:917–931
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J (2012) Introducing EzTaxon–e: a prokaryotic 16S rRNA Gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721
Kimura M (1983) The neutral theory of molecular evolution. Cambridge University Press, Cambridge
Kumar S, Dudley J, Nei M, Tamura K (2008) MEGA: a biologist–centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform 9:299–306
La Scola B, Gundi VA, Khamis A, Raoult D (2006) Sequencing of the rpoB gene and flanking spacers for molecular identification of Acinetobacter species. J Clin Microbiol 44:827–832
Li W, Zhang D, Huang X, Qin W (2014a) Acinetobacter harbinensis sp. nov., isolated from river water. Int J Syst Bacteriol 64(5):1507–1513
Li Y, He W, Wang T, Piao CG, Guo LM, Chang JP, Xie SJ (2014b) Acinetobacter qingfengensis sp. nov., isolated from canker bark of Populus × euramericana. Int J Syst Evol Microbiol 64:1043–1050
Mesbah M, Premachandran U, Whitman WB (1989) Precise measurement of the G+C content of deoxyribonucleic acid by high–performance liquid chromatography. Int J Syst Bacteriol 39:159–167
Minnikin DE, Patel PV, Alshamaony L, Goodfellow M (1977) Polar lipid composition in the classification of Nocardia and related bacteria. Int J Syst Bacteriol 27:104–117
Minnikin DE, O’Donnel AG, Goodfellow M, Alderson G, Athalye M, Schaal A, Parleet JH (1984) An integrated procedure for the extraction of bacterial isoprenoid quinines and polar lipids. J Microbiol Methods 2:233–241
Moore DD, Dowhan D (1995) Preparation and analysis of DNA. In: Ausubel FW, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (eds) Current protocols in molecular biology. Wiley, New York, pp 2–11
Nemec A, Dijkshoorn L, Cleenwerck I, De Baere T, Janssens D, Van Der Reijden TJK, Jezek P, Vaneechoutte M (2003) Acinetobacter parvus sp. nov., a small–colony–forming species isolated from human clinical specimens. Int J Syst Evol Microbiol 53:1563–1567
Nemec A, Musílek M, Maixnerová M, De Baere T, van der Reijden TJK, Vaneechoutte M, Dijkshoorn L (2009) Acinetobacter beijerinckii sp. nov. and Acinetobacter gyllenbergii sp. nov., haemolytic organisms isolated from humans. Int J Syst Evol Microbiol 59:118–124
Nemec A, Musílek M, Šedo O, De Baere T, Maixnerová M, van der Reijden TJ, Dijkshoorn L (2010) Acinetobacter bereziniae sp. nov. and Acinetobacter guillouiae sp. nov., to accommodate Acinetobacter genomic species 10 and 11, respectively. Int J Syst Evol Microbiol 60:896–903
Nemec A, Krizova L, Maixnerova M, Sedo O, Brisse S, Higgins PG (2015) Acinetobacter seifertii sp. nov., a member of the Acinetobacter calcoaceticus–Acinetobacter baumannii complex isolated from human clinical specimens. Int J Syst Bacteriol ijs: 0.000043
Nishimura Y, Ino T, Iizuka H (1988) Acinetobacter radioresistens sp. nov. isolated from cotton and soil. Int J Syst Bacteriol 38:209–211
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Bio Evol 4:406–425
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. MIDI Technical Note 101. MIDI Inc, Newark
Stackebrandt E, Goebel BM (1994) Taxonomic note: a place for DNA–DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Evol Microbiol 44:846–849
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Tindall BJ (1990) Lipid composition of Halobacterium lacusprofundi. FEMS Microbiol Lett 66:199–202
Vaneechoutte M, De Baere T, Nemec A, Musílek M, van der Reijden TJ, Dijkshoorn L (2008) Reclassification of Acinetobacter grimontii Carr et al. 2003 as a later synonym of Acinetobacter junii Bouvet and Grimont 1986. Int J Syst Evol Microbiol 58:937–940
Vaz-Moreira I, Novo A, Hantsis-Zacharov E, Lopes AR, Gomila M, Nunes OC, Manaia CM, Halpern M (2011) Acinetobacter rudis sp. nov., isolated from raw milk and raw wastewater. Int J Syst Evol Microbiol 61:2837–2843
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703
Acknowledgments
This work was conducted under the industrial infrastructure program (No. N0000888) for fundamental technologies which is funded by the Ministry of Trade, Industry & Energy (MOTIE, Korea) and The High-end Foreign Expert Project of China, No. GDT20152100019.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Erko Stackebrandt.
The NCBI GenBank accession numbers for the 16S rRNA gene sequence and rpoB gene sequence of strain THG–SQM11T are KM598254 and KR856237, respectively.
Electronic supplementary material
Below is the link to the electronic supplementary material.
203_2016_1199_MOESM1_ESM.pdf
Supplementary Fig. S1. Maximum-likelihood tree based on 16S rRNA gene sequences showing the relationships between strain THG–SQM11T and other related type species. Numbers at nodes (over 50% are shown) represent percentages of bootstrap support based on a maximum-likelihood analysis of 1,000 resampled datasets. Moraxella lacunata ATCC 17967T (AF005160) was used as outgroup (PDF 13 kb)
203_2016_1199_MOESM2_ESM.pdf
Supplementary Fig. S2. Neighbour-joining phylogenetic tree based on rpoB gene sequences showing the relationships of strain THG–SQM11T with related Acinetobacter species. Bootstrap values (expressed as percentage of 1,000 replications) over 70% are shown at branch points. Bar, 0.02 substitutions per nucleotide position (PDF 15 kb)
203_2016_1199_MOESM3_ESM.pdf
Supplementary Fig. S3. Transmission electron micrograph of cells of THG–SQM11T. The detection was performed after negative staining with uranyl acetate. Bar, 0.5µm (PDF 128 kb)
203_2016_1199_MOESM4_ESM.pdf
Supplementary Fig. S4. Two-dimensional thin-layer chromatography of polar lipids of strain THG–SQM11T (a1, a2, and a3) and A. junii KCTC 12416T (b1, b2, and b3). a1 and b1: Total lipids detected by spraying with 5 % molybdophosphoric acid; a2 and b2: aminolipids detected by spraying with 0.2 % ninhydrin; a3 and b3: phospholipids detected by spraying with molybdenum blue. Abbreviations: Diphosphatidylglycerol (DPG), phosphatidylglycerol (PG), phosphatidylethanolamine (PE), phosphatidylcholine (PC), unidentified aminolipid (AL), unidentified lipid (L) (PDF 120 kb)
Rights and permissions
About this article
Cite this article
Du, J., Singh, H., Yu, H. et al. Acinetobacter plantarum sp. nov. isolated from wheat seedlings plant. Arch Microbiol 198, 393–398 (2016). https://doi.org/10.1007/s00203-016-1199-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-016-1199-3