Abstract
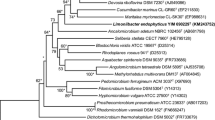
A novel Gram-staining negative, motile, rod-shaped and aerobic bacterial strain, designated EGI 60010T, was isolated from healthy roots of Glycyrrhiza uralensis F. collected from Yili County, Xinjiang Province, North-West China. The 16S rRNA gene sequence of strain EGI 60010T showed 97.2 % sequence similarities with Ochrobactrum anthropi ATCC 49188T and Ochrobactrum cytisi ESC1T, and 97.1 % with Ochrobactrum lupini LUP21T. The phylogenetic analysis based on 16S rRNA gene sequences showed that the new isolate clustered with members of the genera Ochrobactrum, and formed a distinct clade in the neighbour-joining tree. Q-10 was identified as the respiratory quinone for strain EGI 60010T. The major fatty acids were summed feature 8 (C18:1 ω6c and/or C18:1 ω7c), C19:0 cyclo ω8c, summed feature 4 (C17:1 iso I/anteiso B) and C16:0. The polar lipids detected were diphosphatidylglycerol, phosphatidylethanolamine, phosphatidylmethylethanolamine, phosphatidylglycerol and phosphatidylcholine. The DNA G+C content of strain EGI 60010T was determined to be 60.4 mol%. The genomic DNA relatedness values determined between strain EGI 60010T and the closely related strains O. anthropi JCM 21032T, O. cytisi CCTCC AB2014258T and O. lupini NBRC 102587T were 50.3, 50.0 and 41.6 %, respectively. Based on the results of the molecular studies supported by its differentiating phenotypic characteristics, strain EGI 60010T was considered to represent a novel species within the genus Ochrobactrum, for which the name Ochrobactrum endophyticum sp. nov., is proposed. The type strain is EGI 60010T (=CGMCC 1.15082T = KCTC 42485T = DSM 29930T).


Similar content being viewed by others
References
Cappuccino JG, Sherman N (2002) Microbiology: a laboratory manual, 6th edn. Benjamin/Cummings, Menlo Park
Cerny G (1978) Studies on aminopeptidase for the distinction of Gram-negative from Gram-positive bacteria. Appl Microbiol Biotechnol 5:113–122
Christensen H, Angen O, Mutters R, Olsen JE, Bisgaard M (2000) DNA–DNA hybridization determined in microwells using covalent attachment of DNA. Int J Syst Evol Microbiol 50:1095–1102
Collins MD, Jones D (1980) Lipids in the classification and identification of coryneform bacteria containing peptidoglycan based on 2,4-diaminobutyric acid. Appl Bacteriol 48:459–470
Collins MD, Pirouz T, Goodfellow M, Minnikin DE (1977) Distribution of menaquinones in actinomycetes and corynebacteria. J Gen Microbiol 100:221–230
Ezaki T, Hashimoto Y, Yabuuchi E (1989) Fluorometric deoxyribonucleic acid-deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int J Syst Bacteriol 39:224–229
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Syst Zool 20:406–416
He L, Li W, Huang Y, Wang LM, Liu ZH, Lanoot BJ, Vancanneyt M, Swings J (2005) Streptomyces jietaisiensis sp. nov., isolated from soil in northern China. Int J Syst Evol Microbiol 55:1939–1944
Holmes B, Popoff M, Kiredjian M, Kersters K (1988) Ochrobactrum anthropi gen. nov., sp. nov. from human clinical specimens and previously known as group Vd. Int J Syst Bacteriol 38:406–416
Huber B, Scholz HC, Kámpfer P, Falsen E, Langer S, Busse HJ (2010) Ochrobactrum pituitosum sp. nov., isolated from an industrial environment. Int J Syst Evol Microbiol 60:321–326
Imran A, Hafeez FY, Frühling A, Schumann P, Malik KA, Stackebrandt E (2010) Ochrobactrum ciceri sp. nov., isolated from nodules of Cicer arietinum. Int J Syst Evol Microbiol 60:1548–1553
Kämpfer P, Buczolits S, Albrecht A, Busse HJ, Stackebrandt E (2003) Towards a standardized format for the description of a novel species (of an established genus): Ochrobactrum gallinifaecis sp. nov. Int J Syst Evol Microbiol 53:893–896
Kämpfer P, Scholz HC, Huber B, Falsen E, Busse HJ (2007) Ochrobactrum haematophilum sp. nov. and Ochrobactrum pseudogrignonense sp. nov., isolated from human clinical specimens. Int J Syst Evol Microbiol 57:2513–2518
Kämpfer P, Sessitsch A, Schloter M, Huber B, Busse HJ, Scholz HC (2008) Ochrobactrum rhizosphaerae sp. nov. and Ochrobactrum thiophenivorans sp. nov., isolated from the environment. Int J Syst Evol Microbiol 58:1426–1431
Kämpfer P, Huber B, Busse HJ, Scholz HC, Tomaso H, Hotzel H, Melzer F (2011) Ochrobactrum pecoris sp. nov., isolated from farm animals. Int J Syst Evol Microbiol 61:2278–2283
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon Y, Lee JH, Yi H, Won S, Chun J (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Kroppenstedt RM (1982) Separation of bacterial menaquinones by HPLC using reverse phase (RP18) and a silver loaded ion exchanger as stationary phases. J Liq Chromatogr 5:2359–2387
Lebuhn M, Achouak W, Schloter M, Berge O, Meier H, Barakat M, Hartmann A, Heulin T (2000) Taxonomic characterization of Ochrobactrum sp. isolates from soil samples and wheat roots, and description of Ochrobactrum tritici sp. nov. and Ochrobactrum grignonense sp. nov. Int J Syst Evol Microbiol 50:2207–2223
Leifson E (1960) Atlas of bacterial flagellation. Academic Press, London
Li WJ, Xu P, Schumann P, Zhang YQ, Pukall R, Xu LH, Stackebrandt E, Jiang CL (2007) Georgenia ruanii sp. nov., a novel actinobacterium isolated from forest soil in Yunnan (China), and emended description of the genus Georgenia. Int J Syst Evol Microbiol 57:1424–1428
Mesbah M, Premachandran U, Whitman WB (1989) Precise measurement of the G+C content of deoxyribonucleic acid by high performance liquid chromatography. Int J Syst Bacteriol 39:159–167
Ming H, Nie GX, Jiang HC, Yu TT, Zhou EM, Feng HG, Tang SK, Li WJ (2012) Paenibacillus frigoriresistens sp. nov., a novel psychrotroph isolated from a peat bog in Heilongjiang, Northern China. Antonie Van Leeuwenhoek 102:297–305
Minnikin DE, Collins MD, Goodfellow M (1979) Fatty acid and polar lipid composition in the classification of Cellulomonas, Oerskovia and related taxa. J Appl Bacteriol 47:87–95
Parte AC (2014) LPSN—list of prokaryotic names with standing in nomenclature. Nucleic Acids Res 42(D1):D613–D616
Qin S, Li J, Chen HH, Zhao GZ, Zhu WY, Jiang CL, Xu LH, Li WJ (2009) Isolation, diversity, and antimicrobial activity of rare actinobacteria from medicinal plants of tropical rain forests in Xishuangbanna, China. Appl Environ Microbiol 75:6176–6186
Reddy GSN, Garcia-Pichel F (2005) Dyadobacter crusticola sp. nov., from Biological Soil Crusts in the Colorado Plateau, USA and emended description of the genus Dyadobacter Chelius and Tripplett 2000. Int J Syst Evol Microbiol 55:1295–1299
Reddy GSN, Nagy M, Garcia-Pichel F (2006) Belnapia moabensisgen nov., sp. nov., an a proteobacterium from biological soil crusts in the Colorado Plateau, USA. Int J SystEvol Microbiol 56:51–58
Rivas R, Velázquez E, Palomo J, Mateos PF, García-Benavides P, Martínez-Molina E (2002) Rapid identification of Clavibacter michiganensis subspecies sepedonicus using two primers random amplified polymorphic DNA (TP-RAPD) fingerprints. Eur J Plant Pathol 108:179–184
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. MIDI Technical Note 101. MIDI Inc., Newark
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Teyssier C, Marchandin H, Jean-Pierre H, Masnou A, Dusart G, Jumas-Bilak E (2007) Ochrobactrum pseudintermedium sp. nov., a novel member of the family Brucellaceae, isolated from human clinical samples. Int J Syst Evol Microbiol 57:1007–1013
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Tripathi AK, Verma SC, Chowdhury SP, Lebuhn M, Gattinger A, Schloter M (2006) Ochrobactrum oryzae sp. nov., an endophytic bacterial species isolated from deep-water rice in India. Int J Syst Evol Microbiol 56:1677–1680
Trujillo ME, Willems A, Abril A, Planchuelo AM, Rivas R, Ludeña D, Mateos PF, Martinez-Molina E, Velázquez E (2005) Nodulation of Lupinus albus by strains of Ochrobactrum lupini sp. nov. Appl Environ Microbiol 71:1318–1327
Velasco J, Romero C, López-Goñi I, Leiva J, Díaz R, Moriyón I (1998) Evaluation of the relatedness of Brucella spp. and Ochrobactrum anthropi and description of Ochrobactrum intermedium and description of Ochrobactrum intermedium sp. nov., a new species with a closer relationship to Brucella spp. Int J Syst Bacteriol 48:759–768
Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, Moore LH, Moore WEC, Murray RGE, Stackebrandt E, Starr MP, Trüper HG (1987) International committee on systematic bacteriology. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol 37:463–464
Woo SG, Ten LN, Park J, Lee J (2011) Ochrobactrum daejeonense sp. nov., a nitrate-reducing bacterium isolated from sludge of a leachate treatment plant. Int J Syst Evol Microbiol 61:2690–2696
Xu P, Li WJ, Tang SK, Zhang YQ, Chen GZ, Chen HH, Xu LH, Jiang CL (2005) Naxibacter alkalitolerans gen. nov., sp. nov., a novel member of the family ‘Oxalobacteraceae’ isolated from China. Int J Syst Evol Microbiol 55:1149–1153
Zurdo-Piñeiro JL, Rivas R, Trujillo ME, Vizcaíno N, Carrasco JA, Chamber M, Palomares A, Mateos PF, Martínez-Molina E, Velázquez E (2007) Ochrobactrum cytisi sp. nov. isolated from nodules of Cytisus scoparius in Spain. Int J Syst Evol Microbiol 57:784–788
Acknowledgments
The authors are grateful to Prof. Dr. Takuji Kudo (JCM, Japan), Dr. Tomohiko Tamura (NBRC, Japan) and Dr. Fang Peng (CCTCC, China) for kindly providing the reference type strains. This research was supported by the National Natural Science Foundation of China (No. 31200008), West Light Foundation of the Chinese Academy of Sciences (XBBS201305), Projects of China Tobacco Yunnan Industrial Co. Ltd. (Nos. 2014YL01 and 2015CP01), the Hundred Talents Program of Chinese Academy of Sciences and the High-level Talents Program of Xinjiang Autonomous Region. W-J Li was also supported by Guangdong Province Higher Vocational Colleges and Schools Pearl River Scholar Funded Scheme (2014).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Communicated by Erko Stackebrandt.
Li Li and Yan-Qiong Li have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, L., Li, YQ., Jiang, Z. et al. Ochrobactrum endophyticum sp. nov., isolated from roots of Glycyrrhiza uralensis . Arch Microbiol 198, 171–179 (2016). https://doi.org/10.1007/s00203-015-1170-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-015-1170-8