Abstract
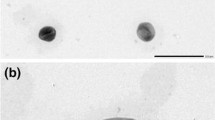
A Gram-negative, non-mobile, polar single flagellum, rod-shaped bacterium WZBFD3-5A2T was isolated from a wheat soil subjected to herbicides for several years. Cells of strain WZBFD3-5A2T grow optimally on Luria-Bertani agar medium at 30 °C in the presence of 0–4.0 % (w/v) NaCl and pH 8.0. 16S rRNA gene sequence analysis revealed that strain WZBFD3-5A2T belongs to the genus Pseudomonas. Physiological and biochemical tests supported the phylogenetic affiliation. Strain WZBFD3-5A2T is closely related to Pseudomonas nitroreducens IAM1439T, sharing 99.7 % sequence similarity. DNA–DNA hybridization experiments between the two strains showed only moderate reassociation similarity (33.92 ± 1.0 %). The DNA G+C content is 62.0 mol%. The predominant respiratory quinine is Q-9. The major cellular fatty acids present are C16:0 (28.55 %), C16:1ω6c or C16:1ω7c (20.94 %), C18:1ω7c (17.21 %) and C18:0 (13.73 %). The isolate is distinguishable from other related members of the genus Pseudomonas on the basis of phenotypic and biochemical characteristics. From the genotypic, chemotaxonomic and phenotypic data, it is evident that strain WZBFD3-5A2T represents a novel species of the genus Pseudomonas, for which the name Pseudomonas nitritereducens sp. nov. is proposed. The type strain is WZBFD3-5A2T (=CGMCC 1.10702T = LMG 25966T).


Similar content being viewed by others
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Andrews JM (2008) BSAC standardized disc susceptibility testing method (Version 7). J Antimicrob Chemother 62:256–278
Cappuccino JG, Sherman N (2002) Microbiology: a Laboratory Manual, 6th edn. Pearson Education, Inc. and Benjamin Cummings, San Francisco
Chun J, Lee JH, Jung Y, Kim M, Kim S, Kim BK, Lim YW (2007) EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int J Syst Evol Microbiol 57:2259–2261
Clarke PH (1953) Hydrogen sulphide production by bacteria. J Gen Microbiol 8:397–407
De Ley J, Cattoir H, Reynaerts A (1970) The quantitative measurement of DNA hybridization from renaturation rates. Eur J Biochem 12:133–142
Denner EB, Paukner S, Kampfer P, Moore ER, Abraham WR, Busse HJ, Wanner G, Lubitz W (2001) Description of Sphingomonas pituitosa sp. nov., a novel polysaccharide-producing sphingomonad. Int J Syst Evol Microbiol 51:827–841
Dong XZ, Cai MY (2001) Determinative manual for routine bacteriology. Scientific Press, Beijing (in Chinese)
Embley TM (1991) The linear PCR reaction: a simple and robust method for sequencing amplified rRNA genes. Lett Appl Microbiol 13:171–174
Fraser SL, Jorgensen JH (1997) Reappraisal of the antimicrobial susceptibilities of Chtyseobacterium and Flavobacterium species and methods for reliable susceptibility testing. Antimicrob Agents Chemother 41:2738–2741
Gerhardt P, Murray RGE, Wood WA, Krieg NR (1994) Methods for General and Molecular Bacteriology. American Society for Microbiology, Washington, DC
Gupta SK, Kumari R, Prakash O, Lal R (2008) Pseudomonas panipatensis sp. nov., isolated from an oil-contaminated site. Int J Syst Evol Microbiol 58:1339–1345
Hiraishi A, Ueda Y, Ishihara J (1998) Quinone profiling of bacterial communities in natural and synthetic sewage activated sludge for enhanced phosphate removal. Appl Environ Microbiol 64:992–998
Johnson JL (1994) Similarity analysis of DNAs. In: Gerhardt PE, Murray RG, Wood WA, Krieg NR (eds) Methods for general and molecular bacteriology. American Society for Microbiology, Washington, pp 655–681
Kwon SW, Kim JS, Park IC, Yoon SH, Park DH, Lim CK, Go SJ (2003) Pseudomonas koreensis sp. nov., Pseudomonas umsongensis sp. nov. and Pseudomonas jinjuensis sp. nov., novel species from farm soils in Korea. Int J Syst Evol Microbiol 53:21–27
Lang E, Griese B, Spröer C, Schumann P, Steffen M, Verbarg S (2007) Characterization of ‘Pseudomonas azelaica’ DSM 9128, leading to emended descriptions of Pseudomonas citronellolis Seubert 1960 (Approved Lists 1980) and Pseudomonas nitroreducens Iizuka and Komagata 1964 (Approved Lists 1980), including Pseudomonas multiresinivorans as its later heterotypic synonym. Int J Syst Evol Microbiol 57:878–882
Las Heras A, Domínguez L, López I, Fernández-Garayzábal JF (1999) Outbreak of acute ovine mastitis associated with Pseudomonas aeruginosa infection. Vet Rec 145:111–112
Liu M, Luo XS, Zhang L, Dai J, Wang Y, Tang YL, Li J, Sun TJ, Fang CX (2009) Pseudomonas xinjiangensis sp. nov., a moderately thermotolerant bacterium isolated from desert sand. Int J Syst Evol Microbiol 59:1286–1289
Mandel M, Igambi L, Bergenda J, Dodson ML, Scheltge E (1970) Correlation of melting temperature and cesium chloride buoyant density of bacterial deoxyribonucleic acid. J Bacteriol 101:333–338
Marmur J (1961) A procedure for the isolation of deoxyribonucleic acid from microorganisms. J Mol Biol 3:208–218
Mata JA, Martínez-Cánovas J, Quesada E, Béjar V (2002) A detailed phenotypic characterisation of the type strains of Halomonas species. Syst Appl Microbiol 25:360–375
MIDI (2005) Sherlock Microbial Identification System Operating Manual, version 6.0. Newark DE: MIDI Inc
Migula W (1894) Uber ein neues System der Bakterien. Arb Bakteriol Inst Karlsruhe 1:235–238
Palleroni NJ (1984) Genus I. Pseudomonas Migula 1894, 237AL (nom. cons. Opin. 5, Jud. Comm. 1952, 237). In: Krieg NR, Holt JG (eds) Bergey’s Manual of Systematic Bacteriology, vol. 1. Williams & Wilkins, Baltimore, pp 141–199
Prakash O, Kumari K, Lal R (2007) Pseudomonas delhiensis sp. nov., from a fly ash dumping site of a thermal power plant. Int J Syst Evol Microbiol 57:527–531
Qin QL, Zhao DL, Wang J, Chen XL, Dang HY, Li TG, Zhang YZ, Gao PJ (2007) Wangia profunda gen. nov., sp. nov., a novel marine bacterium of the family Flavobacteriaceae isolated from southern Okinawa trough deep-sea sediment. FEMS Microbiol Lett 271:53–58
Romano I, Lama L, Nicolaus B, Poli A, Gambacorta A, Giordano A (2006) Halomonas alkaliphila sp. nov., a novel halotolerant alkaliphilic bacterium isolated from a salt pool in Campania (Italy). J Gen Appl Microbiol 52:339–348
Stanier RY, Palleroni NJ, Doudoroff M (1966) The aerobicpseudomonads: a taxonomic study. J Gen Microbiol 43:159–271
Stolz A, Busse HJ, Kämpfer P (2007) Pseudomonas knackmussii sp. nov. Int J Syst Evol Microbiol 57:572–576
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Wang YN, Cai H, Yu SL, Wang ZY, Wu XL (2007) Halomonas gudaonensis sp. nov., isolated from a saline soil contaminated by crude oil. Int J Syst Evol Microbiol 57:911–915
Wu JF, Shen XH, Zhou YG, Liu SJ (2004) Characterization of p-chloronitrobenzene-degrading Comamonas sp. CNB1 and its degradation of p-chloronitrobenzene. Acta Microbiol Sinica 44:8–12
Zumft WG (1992) The denitrifying bacteria. In: Balows A, Truper HG, Dworkin M, Harder W, Schleifer KH (eds) The Prokaryotes, 2nd edn. Springer, New York, pp 554–582
Acknowledgments
The authors thank Professor De-Rong An (Shanxi Key Laboratory of Molecular Biology for Agriculture, College of Plant Protection, Northwest Agricultural & Forestry University, Yanglin, China) for providing the transmission electron microscopy facility and Professor Xiao-Jun Yan (Ningbo University, China) for excellent technical assistance. This work was supported by the Knowledge Innovation Program of the Chinese Academy of Sciences (Grant: KSCX2-YW-G-052) and by the National Natural Science Foundation of China (Grant: 31100582).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Erko Stackebrandt.
Ya-Nan Wang and Wei-Hong He contributed equally to this work.
Sequence deposited: The GenBank accession number for the 16S rRNA gene sequence of strain WZBFD3-5A2T is HM246143.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, YN., He, WH., He, H. et al. Pseudomonas nitritireducens sp. nov., a nitrite reduction bacterium isolated from wheat soil. Arch Microbiol 194, 809–813 (2012). https://doi.org/10.1007/s00203-012-0838-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-012-0838-6