Abstract
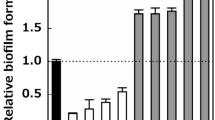
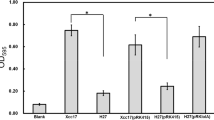
Biofilm formation mutants are often found to have defective or altered motility. The motility phenotype was exploited to identify Pseudomonas fluorescens biofilm formation mutants. Fourteen motility mutants were obtained from P. fluorescens isolate TC222 and eight stable mutants were studied further. The eight transposon insertion mutants showed altered ability to form biofilm compared with the parent. Five Tn5-inserted genes from these mutants were cloned and sequenced. Genetic analysis showed that two insertions were located in genes affecting multiple cell surface characteristics, including lipopolysaccharide (rfbD) and polar flagella (fliR). Three genes encoding for a putative Mig-14 family protein (epsB), a probable bacteriophage signal peptide protein (bspA) and a soluble pyridine nucleotide transhydrogenase (pyrA) were reported for the first time to be involved in biofilm formation. Complementation experiments of rfbD and epsB genes proved that biofilm formation of the corresponding mutants could be restored. Further semi-quantitative reverse transcription-PCR analysis showed that both rfbD and epsB can express their transcripts much higher in the complemented strains than that in wild-type strains. The transcripts of both genes in their mutants could not be detected.





Similar content being viewed by others
Abbreviations
- LPS:
-
Lipopolysaccharide
- NBY:
-
Nutrient broth containing yeast extract medium
- ORFs:
-
Open reading frames
- STH:
-
Soluble pyridine nucleotide transhydrogenase
- SDS:
-
Sodium dodecylsulfate
- SQRT-PCR:
-
Semi-quantitative reverse transcription-PCR
References
Andersen JB, Sternberg C, Poulsen LK, Bjørn SP, Givskov M, Molin S (1998) New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl Environ Microbiol 64:224–2246
Brodsky IE, Ghori N, Falkow S, Monack D (2005) Mig-14 is an inner membrane-associated protein that promotes Salmonella typhimurium resistance to CRAMP, survival within activated macrophages and persistent infection. Mol Microbiol 55:954–972
Caiazza NC, O’Toole GA (2004) SadB is required for the transition from reversible to irreversible attachment during biofilm formation by Pseudomonas aeruginosa PA14. J Bacteriol 186:4476–4485
Costerton JW, Lewandowski Z, Caldwell DE, Korver DR, Lappin-Scott HM (1995) Microbial biofilms. Annu Rev Microbiol 15:711–745
Danese PN, Pratt LA, Kolter R (2000) Exopolysaccharide production is required for development of the Escherichia coli K-12 biofilm architecture. J Bacteriol 68:1967–1974
Davies DG, Chakrabarty AM, Geesey GG (1993) Exopolysaccharide production in biofilms: substratum activation of alginate gene expression by Pseudomonas aeruginosa. Appl Environ Microbiol 59:1181–1186
Dekkers LC, Van der Bij AJ, Mulders IH, Phoelich CC, Wentwood RA, Glandorf DC, Wijffelman CA, Lugtenberg BJ (1998) Role of the O-antigen of lipopolysaccharide, and possible roles of growth rate and of NADH:ubiquinone oxidoreductase (nuo) in competitive tomato root-tip colonization by Pseudomonas fluorescens WCS365. Mol Plant Microbe Interact 11:763–771
Duffy BR, Defago G (2000) Controlling instability in gacS-gacA regulatory genes during inoculant production of Pseudomonas fluorescens biocontrol strains. Appl Environ Microbiol 66:3142–3150
Fletcher M (1988) Attachment of Pseudomonas fluorescens to glass and influence of electrolytes on bacterium substratum separation distance. J Bacteriol 170:2027–2030
Fraser GM, Hirano T, Ferris HU, Devgan LL, Kihara M, Macnab RM (2003) Substrate specificity of type III flagellar protein export in Salmonella is controlled by subdomain interactions in FlhB. Mol Microbiol 48:1043–1057
French CE, Boonstra B, Bufton AJ, Bruce NC (1997) Cloning, sequence, and properties of the soluble pyridine nucleotide transhydrogenase of Pseudomonas fluorescens. J Bacteriol 179:2761–765
Gallegos MT, Schleif RA, Bairoch K, Hofmann K, Ramos JL (1997) Arac/XylS family of transcriptional regulators. Microbiol Mol Biol Rev 61:393–410
Genevaux P, Bauda P, Michael S, Oudega DB (1999) Identification of Tn-10 insertions in the rfaG, rfaP, and galU genes involved in lipopolysaccharide core biosynthesis that affect Escherichia coli adhesion. Arch Microbiol 172:1–8
Hitchcock PJ, Brown TM (1983) Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J Bacteriol 154:269–277
Hu YQ, Zhang J, Song FP, Shu CL, Huang DF (2004) Construction of stable shuttle expression vectors pQMV and pQMV (in Chinese with English abstract). J Agric Biotechnol 12:202–205
Kim W, Surette MG (2003) Swarming populations of Salmonella represent a unique physiological state coupled to multiple mechanisms of antibiotic resistance. Biol Proced 5:189–196
Klausen M, Heydorn A, Ragas P, Lambertsen L, Aaes-Jorgensen A, Molin S, Tolker-Nielsen T (2003) Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol Microbiol 48:1511–1524
Korber DR, James GA, Costerton JW (1994) Evaluation of fleroxacin activity against established Pseudomonas fluorescens Biofilms. Appl Environ Microbiol 60(5):1663–1669
Landini P, Zehnder AJ (2002) The global regulatory hns gene negatively affects adhesion to solid surfaces by anaerobically grown Escherichia coli by modulating expression of flagellar genes and lipopolysaccharide production. J Bacteriol 184:1522–1529
Lopez-Lopez G, Pascual A, Perea J (1991) Effect of plastic catheter material on bacterial adherence and viability. J Med Microbiol 34:349–353
Mah TF, O’Toole GA (2001) Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol 9:34–39
Martin RG, Rosner JL (2001) The AraC transcriptional activators. Curr Opin Microbiol 4:132–137
Miller J (1972) Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Nian HJ, Zhang J, Fan LQ, Liu S, Song FP, Huang DF (2007) Application of ARDRA technology in isolation and identification of Pseudomonas fluorescens. Agri Sci in China 40(1):92–98
Ohmura N, Kitamura K, Saiki H (1993) Selective adhesion of Thiobacillus ferroodoxidans to pyrite. Appl Environ Microbiol 59:4044–4050
O’Toole GA, Kolter R (1998a) Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signaling pathways: a genetic analysis. Mol Microbiol 28:449–461
O’Toole GA, Kolter R (1998b) Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol 30:295–304
Pardee AB, Jacob F, Monod J (1959) The genetic control and cytoplasmic expression of “inducibility” in the synthesis of galactosidase in E. coli J Mol Biol 1:165–178
Piette JP, Idziak ES (1992) A model study of factors involved in adhesion of Pseudomonas fluorescens to meat. Appl Environ Microbiol 58:2783–2791
Potera C (1996) Research news: biofilms invade microbiology. Science 273:1795–1797
Pratt LA, Kolter R (1998) Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol 30:285–293
Rahim R, Burrows LL, Monteiro MA, Perry MB, Lam JS (2000) Involvement of the rml locus in core oligosaccharide and O polysaccharide assembly in Pseudomonas aeruginosa. Microbiology 146:2803–2814
Ramphal R, Koo L, Ishimoto KS, Totten PA, Lara JC, Lory S (1991). Adhesion of Pseudomonas aeruginosa pilin deficient mutants to mucin. Infect Immun 59:1307–1311
Reid G, Khoury AE, Preston CA, Costerton JW (1994) Influence of dextrose dialysis solutions on the adhesion of Staphyloccocus aureus and Pseudomonas aeruginosa to three catheter surfaces. Am J Nephrol 14:37–40
Sambrook J, Fretsh EF, Maniatis T (1989) Molecular cloning: a Laboratory Manual, 2nd edn. Cold Spring Harbor Laboratory, Cold Spring Harbor
Sauer K, Camper AK (2001) Characterization of phenotypic changes in Pseudomonas putida in response to surface-associated growth. J Bacteriol 183:6579–6589
Scharfman A, Kroczynski H, Carnoy C, Van Brussel E, Lamblin G, Ramphal R, Roussel P (1996) Adhesion of Pseudomonas aeruginosa to respiratory mucins and expression of mucin-binding proteins are increased by limiting iron during growth. Infect Immun 64:5417–5420
Simon R, Preifer U, Ptihler A (1983) A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Biotechnology 1:784–791
Spiers AJ, Rainey PB (2005) The Pseudomonas fluorescens SBW25 wrinkly spreader biofilm requires attachment factor, cellulose fibre and LPS interactions to maintain strength and integrity. Microbiology 151:2829–2839
Stanley PM (1983) Factors affecting the irreversible attachment of Pseudomonas aeruginosa to stainless steel. Can J Mictobiol 29:1493–1499
Sternberg C, Christensen BB, Johansen T, Nielsen AT, Andersen JB, Givskov M, Molin S (1999) Distribution of bacterial growth activity in flow-chamber biofilms. Appl Environ Microbiol 65:4108–4117
Toguchi A, Siano M, Burkart M, Harshey RM (2000) Genetics of swarming motility in Salmonella enterica serovar typhimurium: Critical role for lipopolysaccharide. J Bacteriol 182:6308–6321
Watnick PI, Kolter R (1999) Steps in the development of a Vibrio cholerae biofilm. Mol Microbiol 34:586–595
Watnick PI, Lauriano CM, Klose KE, Croal L, Kolter R (2001) The absence of a flagellum leads to altered colony morphology, biofilm development and virulence in Vibrio cholerae O139. Mol Microbiol 39:223–235
Whiteley M, Bangera MG, Bumgarner RE, Parsek MR, Teitzel GM, Lory S, Greenberg EP (2001) Gene expression in Pseudomonas aeruginosa biofilms. Nature 413:860–864
Williams V, Fletcher M (1996) Pseudomonas fluorescens adhesion and transport through porous media are affected by lipopolysaccharide composition. Appl Environ Microbiol 62:100–104
Acknowledgment
We thank Dr Søren Molin for kindly providing the plasmid pJBA28. This work was supported by National Significant Basic Research Project Program (2003CB114201 and 2001CB109005). Special thanks to Ms. Wendy C Kain (Cornell University) for revising this manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Sebastian Suerbaum.
Rights and permissions
About this article
Cite this article
Nian, H., Zhang, J., Song, F. et al. Isolation of transposon mutants and characterization of genes involved in biofilm formation by Pseudomonas fluorescens TC222. Arch Microbiol 188, 205–213 (2007). https://doi.org/10.1007/s00203-007-0235-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-007-0235-8