Abstract
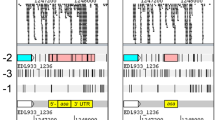
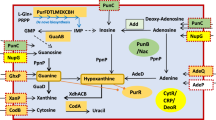
ModE protein, a molybdate sensor/regulator, controls the transcription of genes coding for molybdate uptake (mod), molybdopterin synthesis (moa), molybdoenzymes nitrate reductase (nap) and dimethylsulfoxide reductase (dms), as well as fermentative dihydrogen production (fdhF and hyc) and respiratory nitrate reductase (narXL) in Escherichia coli. The catalytic product of a second protein, MoeA, is also required for molybdate-dependent positive regulation of hyc and nar operons. To explore the potential role of ModE and MoeA in the regulation of other E. coli genes, the global gene expression profile of a wild type and a modE, moeA double mutant grown in glucose-minimal medium under anaerobic conditions were compared. Expression of 67 genes was affected by the modE and moeA mutations (P value <0.01). Of these, 17 differed by at least 2-fold or higher. Fourteen genes were expressed at a higher level in the mutant (2.4- to 23.9-fold) (notably, mod—molybdate transport, deo—nucleoside catabolism and opp—oligopeptide transport operons) and dmsA and yli operon were expressed at a higher level in the wild type parent (2.6- to 5.7-fold). One of the unexpected findings was repression of the deo operon by ModE. This was confirmed by quantitative RT-PCR and by the analysis of a deoC-lacZ fusion. The deo promoter/operator region contains a putative ModE-consensus sequence centered at −35 in which the adenines are replaced by guanines (TGTGT-N7-TGTGT). The ModE protein did bind to the deo upstream DNA and shifted its electrophoretic mobility. Bioinformatics analysis of the E. coli genome for ModE-consensus motif (TATAT-N7-TAYAT) identified 21 additional genes/operons including the moa as potential targets for Mo-control. The physiological role of many of the genes identified solely by bioinformatics (19/21) is unknown. Expression levels of these genes were similar in the parent and the isogenic modE, moeA mutant when cultured anaerobically in glucose-minimal medium. This study identified additional targets, such as deo and opp, for the Mo-dependent control in E. coli.



Similar content being viewed by others
References
Anderson LA, McNairn E, Lubke T, Pau RN, Boxer DH (2000) ModE-dependent molybdate regulation of the molybdenum cofactor operon moa in Escherichia coli. J Bacteriol 182:7035–7043
Bebien M, Kirsch J, Mejean V, Vermeglio A (2002) Involvement of a putative molybdenum enzyme in the reduction of selenate by Escherichia coli. Microbiology 148:3865–3872
Burgis NE, Brucker JJ, Cunningham RP (2003) Repair system for noncanonical purines in Escherichia coli. J Bacteriol 185:3101–3110
Gon S, Patte J-C, Méjean V, Lobbi-Nivol C (2000) The torYZ (yecK bisZ) operon encodes a third respiratory trimethylamine N-oxide reductase in Escherichia coli. J Bacteriol 182:5779–5786
Gonzalez R, Tao H, Shanmugam KT, York SW, Ingram LO (2002) Global gene expression differences associated with changes in glycolytic flux and growth rate in Escherichia coli during the fermentation of glucose and xylose. Biotechnol Prog 18:6–20
Grunden AM, Shanmugam KT (1997) Molybdate transport and regulation in bacteria. Arch Microbiol 168:345–354
Grunden AM, Ray RM, Rosentel JK, Healy FG, Shanmugam KT (1996) Repression of the Escherichia coli modABCD (molybdate transport) operon by ModE. J Bacteriol 178:735–744
Grunden AM, Self WT, Villain M, Blalock JE, Shanmugam KT (1999) An analysis of the binding of repressor protein ModE to modABCD (Molybdate transport) operator/ promoter DNA of Escherichia coli. J Biol Chem 274:24308–24315
Hasona A, Ray RM, Shanmugam KT (1998a) Physiological and genetic analyses leading to identification of a biochemical role for the moeA (molybdate metabolism) gene product in Escherichia coli. J Bacteriol 180:1466–1472
Hasona A, Self WT, Ray RM, Shanmugam KT (1998b) Molybdate-dependent transcription of hyc and nar operons of Escherichia coli requires MoeA protein and ModE-molybdate. FEMS Microbiol Lett 169:111–116
Hille R (1996) The mononuclear molybdenum enzymes. Chem Rev 96:2757–2816
Kisker C, Schindelin H, Rees DC (1997) Molybdenum-cofactor-containing enzymes: structure and mechanism. Ann Rev Biochem 66:233–267
Kisker C, Schindelin H, Baas D, Retey J, Meckenstock RU, Kroneck PM (1999) A structural comparison of molybdenum cofactor-containing enzymes. FEMS Microbiol Rev 22:503–521
Kutsche M, Leimkuhler S, Angermuller S, Klipp W (1996) Promoters controlling expression of the alternative nitrogenase and the molybdenum uptake system in Rhodobacter capsulatus are activated by NtrC, independent of sigma54, and repressed by molybdenum. J Bacteriol 178:2010–2017
Lee JH, Patel P, Sankar P, Shanmugam KT (1985) Isolation and characterization of mutant strains of Escherichia coli altered in H2 metabolism. J Bacteriol 162:344–352
Linn T, St. Pierre R (1990) Improved vector system for constructing transcriptional fusions that ensures independent translation of lacZ. J Bacteriol 172:1077–1084
Lopilato J, Bortner S, Beckwith J (1986) Mutations in a new chromosomal gene of Escherichia coli K-12, pcnB, reduce plasmid copy number of pBR322 and its derivatives. Mol Gen Genet 205:285–290
Maloy S, Nunn WD (1981) Selection for loss of tetracycline resistance by Escherichia coli. J Bacteriol 145:1110–1112
McNicholas PM, Gunsalus RP (2002) The molybdate-responsive Eschrichia coli ModE transcriptional regulator coordinates periplasmic nitrate reductase (napFDAGHBC) operon expression with nitrate and molybdate availability. J Bacteriol 184:3253–3259
McNicholas PM, Chiang RC, Gunsalus RP (1996) The Escherichia coli modE gene: effect of modE mutations on molybdate dependent modA expression. FEMS Microbiol Lett 145:117–123
McNicholas PM, Rech SA, Gunsalus RP (1997) Characterization of the ModE DNA-binding sites in the control regions of modABCD and moaABCDE of Escherichia coli. Mol Microbiol 23:515–524
McNicholas PM, Chiang RC, Gunsalus RP (1998) Anaerobic regulation of the Escherichia coli dmsABC operon requires the molybdate-responsive regulator ModE. Mol Microbiol 27:197–208
Mendel RR, Hansch R (2002) Molybdoenzymes and molybdenum cofactor in plants. J Exp Botany 53:1689–1698
Newman EB, Ahmad D, Walker GC (1982) L-Serine deaminase activity is induced by exposure of Escherichia coli K-12 to DNA-damaging agents. J Bacteriol 152:702–705
Nichols J, Rajagopalan KV (2002) Escherichia coli MoeA and MogA. Function in metal incorporation step of molybdenum cofactor biosynthesis. J Biol Chem 277:24995–25000
Pascal MC, Burini JF, Chippaux M (1984) Regulation of the trimethylamine N-oxide (TMAO) reductase in Escherichia coli: analysis of tor::Mud1 operon fusion. Mol Gen Genet 195:351–355
Pau RN, Klipp W, Leimkuhler S (1997) Molybdenum transport, processing and gene regulation. In: Winkelmann G, Carrano CJ (eds) Transition metals in microbial metabolism. Harwood Academic Publishers, pp 217–234
Rajagopalan KV (1997) Biosynthesis and processing of the molybdenum cofactors. Biochem Soc Trans 25:757–761
Rosentel JK, Healy F, Maupin-Furlow JA, Lee JH, Shanmugam KT (1995) Molybdate and regulation of mod (molybdate transport), fdhF, and hyc (formate hydrogenlyase) operons in Escherichia coli. J Bacteriol 177:4857–4864
Rossmann R, Sawers G, Bock A (1991) Mechanism of regulation of the formate-hydrogenlyase pathway by oxygen, nitrate, and pH: definition of the formate regulon. Mol Microbiol 5:2807–2814
Seeger C, Poulsen C, Dandanell G (1995) Identification and characterization of genes (xapA, xapB, and xapR) involved in xanthosine catabolism in Escherichia coli. J Bacteriol 177:5506–5516
Self WT, Shanmugam KT (2000) Isolation and characterization of mutated Fh1A proteins which activate transcription of the hyc operon (formate hydrogenlyase) of Escherichia coli in the absence of molybdate. FEMS Microbiol Lett 184:47–52
Self WT, Grunden AM, Hasona A, Shanmugam KT (1999) Transcriptional regulation of molybdoenzyme synthesis in Escherichia coli in response to molybdenum: ModE-molybdate, a repressor of the modABCD (molybdate transport) operon is a secondary transcriptional activator for the hyc and nar operons. Microbiology 145:41–55
Self WT, Grunden AM, Hasona A, Shanmugam KT (2001a) Molybdate transport. Res Microbiol 152:311–321
Self WT, Hasona A, Shanmugam KT (2001b) N-terminal truncations in the FhlA protein result in formate- and MoeA- independent expression of the hyc (formate hydrogenlyase) operon of Escherichia coli. Microbiology 147:3093–3104
Self WT, Wolfe MD, Stadtman TC (2003) Cofactor determination and spectroscopic characterization of the selenium-dependent purine hydroxylase from Clostridium purinolyticum. Biochemistry 42:11382–11390
Shin M et al (2001) Repression of deoP2 in Escherichia coli by CytR: conversion of a transcription activator into a repressor. EMBO J 20:5392–5399
Short SA, Singer JT (1984) Studies on deo operon regulation in Escherichia coli: cloning and expression of the deoR structural gene. Gene 31:205–211
Studholme DJ, Pau RN (2003) A DNA element recognised by the molybdenum-responsive transcription factor ModE is conserved in Proteobacteria, green sulphur bacteria and Archaea. BMC Microbiol 3:24–33
Su HS, Lang BF, Newman EB (1989) L-serine degradation in Escherichia coli K-12: cloning and sequencing of the sdaA gene. J Bacteriol 171:5095–5102
Tao H et al (2001) Engineering a homo-ethanol pathway in Escherichia coli: increased glycolytic flux and levels of expression of glycolytic genes during xylose fermentation. J Bacteriol 183:2979–2988
Valentin-Hansen P (1982) Tandem CRP binding sites in the deo operon of Escherichia coli K-12. EMBO J 1:1049–1054
Valentin-Hansen P, Svenningsen BA, Munch-Petersen A, Hammer-Jespersen K (1978) Regulation of the deo operon in Escherichia coli: the double negative control of the deo operon by the cytR and deoR repressors in a DNA directed in vitro system. Mol Gen Genet 159:191–202
Walkenhorst HM, Hemschemeier SK, Eichenlaub R (1995) Molecular analysis of the molybdate uptake operon, modABCD, of Escherichia coli and modR, a regulatory gene. Microbiol Res 150:347–361
Acknowledgements
We thank R. Turnbough and T. Elliott for providing strains and plasmids used in this study. This study was supported by grants from the US Department of Agriculture (00-52104-9704 and 01-35504-10669) and the Department of Energy (DE-FC36-01GO11073 and FG02-96ER20222).
Author information
Authors and Affiliations
Corresponding author
Additional information
Han Tao and Adnan Hasona contributed equally to this work.
Rights and permissions
About this article
Cite this article
Tao, H., Hasona, A., Do, P.M. et al. Global gene expression analysis revealed an unsuspected deo operon under the control of molybdate sensor, ModE protein, in Escherichia coli . Arch Microbiol 184, 225–233 (2005). https://doi.org/10.1007/s00203-005-0039-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-005-0039-7