Abstract
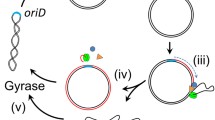
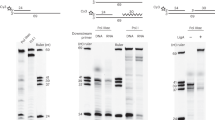
The rolling circle (RC) mechanism of DNA replication generating single-stranded DNA (ssDNA) intermediates is common in various high-copy circular plasmids in Streptomyces, and the ssDNA released after leading strand synthesis is converted to its double-stranded form (dsDNA) by the host proteins. The in vivo and in vitro lagging strand syntheses from ssDNA replicative intermediates of RC plasmid pSN22 in Streptomyces lividans was characterized. The presence or absence of the single-strand origin (sso), the replication initiation site of lagging strand synthesis, did not significantly affect the copy numbers of pSN22 derivatives. In vivo lagging strand synthesis was not affected by the rifampicin inhibition of S. lividans RNA polymerase. Likewise, in vitro lagging strand synthesis using cell-free extracts revealedsso-independent, rifampicin-resistant lagging strand synthesis in S. lividans. Although all four dNTPs are usually required for the initiation of such synthesis, the presence of only one NTP was sufficient to carry outlagging strand synthesis in vitro. Interestingly, the cell-free extract of exponential-phase cells required less ATP than that of stationary-phase cells. These results reveal a predominant RNA polymerase-independent priming system in S. lividans that may be a result of the stabilization of RC plasmids lacking sso in S. lividans.






Similar content being viewed by others
References
Arai K, Kornberg A (1979) A general priming system employing only dnaB protein and primase for DNA replication. Proc Natl Acad Sci USA 76:4308–4312
Arai K, Kornberg A (1981a) Mechanism of dnaB protein action. III. Allosteric role of ATP in the alteration of DNA structure by dnaB protein in priming replication. J Biol Chem 256:5260–5266
Arai K, Kornberg A (1981b) Mechanism of dnaB protein action. IV. General priming of DNA replication by dnaB protein and primase compared with RNA polymerase. J Biol Chem 256:5267–5272
Arai K, Low RL, Kornberg A (1981) Movement and site selection for priming by the primosome in phage ϕX174 DNA replication. Proc Natl Acad Sci USA 78:707–711
Beppu T (1995) Signal transduction and secondary metabolism: prospects for controlling productivity. Trends Biotechnol 13:264–269
Birch P, Khan SA (1992) Replication of single-stranded plasmid pT181 DNA in vitro. Proc Natl Acad Sci USA 89:290–294
Boe L, Gros MF, te Riele H, Ehrlich SD, Gruss A (1989) Replication origins of single-stranded-DNA plasmid pUB110. J Bacteriol 171:3366–3372
Bruand C, Ehrlich SD, Jannière L (1995) Primosome assembly site in Bacillus subtilis. EMBO J 11:2642–2650
Conrad SE, Campbell JL (1979) Characterization of an improved in vitro DNA replication system for Escherichia coli plasmids. Nucl Acids Res 6:3289–3303
Dempsey LA, Zhao AC, Khan SA (1995) Localization of the start sites of lagging-strand replication of rolling-circle plasmids from gram-positive bacteria. Mol Microbiol 15:679–687
Deng ZT, Kieser T, Hopwood DA (1988) “Strong incompatibility” between derivatives of the Streptomyces multi-copy plasmid pIJ101. Mol Gen Genet 214:286–294
Fuller RS, Kaguni JM, Kornberg A (1981) Enzymatic replication of the origin of the Escherichia coli chromosome. Proc Natl Acad Sci USA 78:7370–7374
Gruss A, Ehrlich SD (1989) The family of highly interrelated single-stranded deoxyribonucleic acid plasmids. Microbiol Rev 53:231–241
Gruss A, Ross HF, Novick RP (1987) Functional analysis of a palindromic sequence required for normal replication of several staphylococcal plasmids. Proc Natl Acad Sci USA 84:2165–2169
Hagège J, Pernodet JL, Friedmann A, Guérineau M (1993) Mode and origin of replication of pSAM2, a conjugative integrating element of Streptomyces ambofaciens. Mol Microbiol 10:799–812
Hopwood DA, Bibb MJ, Chater KF, Kieser T, Bruton CJ, Kieser HM, Lydiate DJ, Smith CP, Ward JM, Schrempf H (1985) Genetic manipulation of Streptomyces: a laboratory manual. The John Innes Foundation, Norwich, UK
Inuzuka M, Helinski DR (1978) Replication of antibiotic resistance plasmid R6K DNA in vitro. Biochemistry 17:2567–2573
Jannière L, Gruss A, Ehrlich SD (1993) Plasmids. In: Sonenshein AL, Losick R, Hoch JA (eds) Bacillus subtilis and other gram positive bacteria: biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington DC, pp 625–644
Kaguni JM, Kornberg A (1982) The ρ subunit of RNA polymerase holoenzyme confers specificity in priming M13 viral DNA replication. J Biol Chem 257: 5437–5443
Kataoka M, Seki T, Yoshida, T (1991) Five genes involved in self-transmission of pSN22, a Streptomyces plasmid. J Bacteriol 173:4220–4228
Kataoka M, Kiyose YM, Michisuji Y, Horiguchi T, Seki T, Yoshida T (1994a) Complete nucleotide sequence of the Streptomyces plasmid, pSN22; genetic organization and correlation with genetic properties. Plasmid 32:55–69
Kataoka M, Kuno N, Horiguchi T, Seki T, Yoshida T (1994b) Replication of the Streptomyces plasmid pSN22 through single-stranded intermediates. Mol Gen Genet 242:130–136
Katz E, Thompson CJ, Hopwood, DA (1983) Cloning and expression of the tyrosinase gene from Streptomyces antibioticus in Streptomyces lividans. J Gen Microbiol 129:2703–2714
Khan SA (1997) Rolling-circle replication of bacterial plasmids. Microbiol Mol Biol Rev 61:442–455
Kieser T, Hopwood DA, Wright HM, Thompson CJ (1982) pIJ101, a multi-copy broad host-range Streptomyces plasmid: functional analysis and development of DNA cloning vectors. Mol Gen Genet 185:223–238
Kornberg A, Baker TA (1992) DNA replication, 2nd edn. WH Freeman & Co, New York
Kramer MG, Khan SA, Espinosa M (1997) Plasmid rolling circle replication: identification of the RNA polymerase-directed primer RNA and requirement for DNA polymerase I for lagging strand synthesis. EMBO J. 16:5784–5795
Kramer MG, Espinosa M, Misra TK, Khan SA (1998) Lagging strand replication of rolling-circle plasmids: specific recognition of the ssoA-type origins in different gram-positive bacteria. Proc Natl Acad Sci USA 95:10505–10510
Leenhouts KJ, Tolner B, Bron S, Kok J, Venema G, Seegers JFML (1991) Nucleotide sequence and characterization of the broad-host-range lactococcal plasmid pWVO1. Plasmid 26:55–66
Muth G, Farr M, Hartmann V, Wohlleben W (1995) Streptomyces ghanaensis plasmid pSG5: nucleotide sequence analysis of the self-transmissible minimal replicon and characterization of the replication mode. Plasmid 33:113–126
Novick RP (1989) Staphylococcal plasmids and their replication. Annu Rev Microbiol 43:537–565
Pigac J, Vujaklija D, Toman Z, Gamulin V, Schrempf H (1988) Structural instability of a bifunctional plasmid pZG1 and single-stranded DNA formation in Streptomyces. Plasmid 19:222–230
te Riele H, Michel B, Ehrlich SD (1986a) Single-stranded plasmid DNA in Bacillus subtilis and StaphylococcuStaphylococcus aureus. Proc Natl Acad Sci USA 83:2541–2545
te Riele, H, Michel B, Ehrlich SD (1986b) Are single-stranded circular intermediates involved in plasmid DNA replication? EMBO J 5:631–637
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York
Seegers JFML, Zhao AC, Meijer WJJ, Khan SA, Venema G, Bron S (1995) Structural and functional analysis of the single-strand origin of replication from the lactococcal plasmid pWVO1. Mol Gen Genet 249:43–50
Servín-González L, Sampieri AIII, Cabello J, Galván L, Juárez V, Castro C (1995) Sequence and functional analysis of the Streptomyces phaeochromogenes plasmid pJV1 reveals a modular organization of Streptomyces plasmid that replicate by rolling circle. Microbiol 141:2499–2510
del Solar GH, Moscoso M, Espinosa M. (1993) Rolling-circle replicating plasmids from gram-positive and gram-negative bacteria: a wall falls. Mol Microbiol 8:789–796
Southern EM (1975) Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol 98:503–517
Studier FW, Moffatt BA (1986) Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol 189:113–130
Suzuki I, Kataoka M, Seki T, Yoshida T (1997a) Three single strand origins located on both strands of the Streptomyces rolling circle plasmid pSN22. Plasmid 37:51–64
Suzuki I, Seki T, Yoshida T (1997b) Nucleotide sequence of a nicking site of the Streptomyces plasmid pSN22 replicating by the rolling circle mechanism. FEMS Microbiol Lett 150:283–288
Thompson J, Cundliffe E (1981) Purification and properties of an RNA methylase produced by Streptomyces azureus and involved in resistance to thiostrepton. J Gen Microbiol 124:291–297
Thompson CJ, Ward JM, Hopwood DA (1982) Cloning of antibiotic resistance and nutritional genes in Streptomycetes. J Bacteriol 151:668–677
Thompson J, Rae S, Cundliffe E (1984) Coupled transcription-translation in extracts of Streptomyces lividans. Mol Gen Genet 195:39–43
Yanisch-Perron C, Vieira J, Messing J (1985) Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103–119
Yokoyama E, Doi K, Kimura M, Ogata S (1996) Detection of the single-stranded DNA of Streptomyces plasmid pSA1.1 and a binding histone-like protein. FEMS Microbiol Lett 138:197–200
Zaman S, Radnedge L, Richards H, Ward, JM (1993) Analysis of the site for second-strand initiation during replication of the Streptomyces plasmid pIJ101. J Gen Microbiol 139:669–676
Acknowledgments
We greatly thank to Drs. T. Kieser, H. Araki and T. Ito for valuable discussions; Dr E. Ko-Mitamura for editorial suggestions and manuscript corrections; and Dr H. Masai for valuable comments on the general priming of E. coli.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Suzuki, I., Kataoka, M., Yoshida, T. et al. Lagging strand replication of rolling-circle plasmids in Streptomyces lividans: an RNA polymerase-independent primer synthesis. Arch Microbiol 181, 305–313 (2004). https://doi.org/10.1007/s00203-004-0656-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-004-0656-6