Abstract
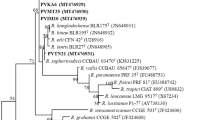
<p>The diversity and taxonomic relationships of 83 bean-nodulating rhizobia indigenous to Ethiopian soils were characterized by PCR-RFLP of the internally transcribed spacer (ITS) region between the 16S and 23S rRNA genes, 16S rRNA gene sequence analysis, multilocus enzyme electrophoresis (MLEE), and amplified fragment-length polymorphism. The isolates fell into 13 distinct genotypes according to PCR-RFLP analysis of the ITS region. Based on MLEE, the majority of these genotypes (70%) was genetically related to the type strain of Rhizobium leguminosarum. However, from analysis of their 16S rRNA genes, the majority was placed with Rhizobium etli. Transfer and recombination of the 16S rRNA gene from presumptively introduced R. etli to local R. leguminosarum is a possible theory to explain these contrasting results. However, it seems unlikely that bean rhizobia originating from the Americas (or Europe) extensively colonized soils of Ethiopia because Rhizobium tropici, Rhizobium gallicum, and Rhizobium giardinii were not detected and only a single ineffective isolate of R. etli that originated from a remote location was identified. Therefore, Ethiopian R. leguminosarum may have acquired the determinants for nodulation of bean from a low number of introduced bean-nodulating rhizobia that either are poor competitors for nodulation of bean or that failed to survive in the Ethiopian environment. Furthermore, it may be concluded from the genetic data presented here that the evidence for separating R. leguminosarum and R. etli into two separate species is inconclusive.





Similar content being viewed by others
References
Amarger N, Macheret V, Laguerre G (1997) Rhizobium gallicum sp. nov. and Rhizobium giardinii sp. nov., from Phaseolus vulgaris nodules. Int J Syst Bacteriol 47:996–1006
Anyango, B, Wilson KJ, Beynon JL, Giller KE (1995) Diversity of Rhizobia nodulating Phaseolus vulgaris L. in two Kenyan Soils with contrasting pHs. Appl Environ Microbiol 61:4016–4021
Beyene D (1985) The state of research on biological nitrogen fixation in Ethiopia. In Ssali H, Keya SO (eds) Biological nitrogen fixation in Africa. Matianum, Nairobi, pp 72–84
Diouf A, de Lajude P, Neyra M, Kersters K, Gillis M, Martinez E, Gueye M (2000) Polyphasic characterization of rhizobia that nodulate Phaseolus vulgaris in West Africa (Senegal and Gambia). Int J Syst Evol Microbiol 50:159–170
Eardly BD, Wang FS, Whittam TS, Selander RK (1995) Species limits in Rhizobium populations that nodulate the common bean (Phaseolus vulgaris). Appl Environ Microbiol 61:507–512
Gepts P (1990) Biochemical evidence bearing on the domestication of Phaseolus (Fabaceae) beans. Econ Bot 44:28–38
Gepts P, Bliss FA (1988) Dissemination pathways of common bean (Phaseolus vulgaris, Fabaceae) deduced from phaseolin electrophoretic variability. II Europe and Africa. Econ Bot 42:86–104
Graham PH, Ballen KG, Montealegre C, Jones RK, Fischer B, Luque E (1999) Characterization of rhizobia associated with Dalea spp. In natural prairies and revegetation areas in Minnesota. In: Martinez E, Hernandez G (eds) Highlights of nitrogen fixation research. Kluwer Academic/Plenum, New York, pp 69–75
Herbert PDN, Beaton MJ (1993) Methodologies for allozyme analysis using cellulose acetate electrophoresis. Department of Zoology, University of Guelph, Guelph, Ontario. Helena Laboratories
Herrera-Cervera JA, Caballero-Mellado J, Laguerre G, Tichy HV, Requena N, Amarger N, Martínez-Romero E, Olivares J, Sanjuán J (1999) At least five rhizobial species nodulate Phaseolus vulgaris in a Spanish soil. FEMS Microbiol Ecol 30:87–97
Jordan DC (1984) Bergey’s manual of systematic bacteriology, vol 1. Williams and Williams, Baltimore, pp 234–256
Kumar S, Tamura K, Nei M (1993) MEGA: Molecular evolutionary genetics analysis, version 1.01. The Pennsylvania State University, University Park, Pennsylvania
Martinez E, Pardo MA, Palacios R, Cevallos MA (1985) Reiteration of nitrogen fixation gene sequences and specificity of Rhizobium in nodulation and nitrogen fixation in Phaseolus vulgaris. J Gen Microbiol 131:1779–1786
Martinez E, Segovia L, Mercante FM, Franco AA, Graham PH, Pardo MA (1991) Rhizobium tropici: a novel species nodulating Phaseolus vulgaris beans and Leucaena sp. trees. Int. J. Syst. Bacteriol. 41:417–426
Pinero D, ER Martinez, RK Selander (1988) Genetic diversity and relationships among isolates of Rhizobium leguminosarum biovar phaseoli. Appl Environ Microbiol 54:2825–2832
Raven PH, Polhill RM (1981) Biogeography of the Leguminosae. In: Polhill RM, Raven PH (eds) Advances in legume systematics. Part 1. Royal Botanic Gardens, Kew, pp 27–34
Rohlf FJ (1988) NTSYS-pc numerical taxonomy and multivariate analysis system. Exeter, Setauket, New York
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York
Segovia L, Young JPW, Martinez-Romero E (1993) Reclassification of American Rhizobium leguminasorum biovar phaseoli type I strains as Rhizobium etli sp. nov. Int J Syst Bacteriol 43:374–377
Selander RK, Caugant DA, Ochman H, Musser JM, Gilmour MN, Whittam TS (1986) Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl Environ Microbiol 51:873–884
Sessitsch A, Ramirez-Saad H, Hardarson G, Akkermans ADL, de Vos WM (1997) Classification of Austrian rhizobia and the Mexican isolate FL27 obtained from Phaseolus vulgaris L. as Rhizobium gallicum. Int J Syst Bacteriol 47:1097–1101
Shimodaira H, Hasegawa M (1999) Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol Biol Evol 16:1114–1116
Sneath PHA, Sokal RR (1973) Numerical taxonomy. W.H. Freeman, San Francisco
Tjahjoleksono, A (1993) Characterisation et diversite des souche de rhizobium nodulant le hurticol (Phaseolus vulgaris) cultive en trios sites tropicaus. PhD thesis. Universite de Lyon
van Berkum P (1990) Evidence for a third uptake hydrogenase phenotype among the soybean bradyrhizobia. Appl Environ Microbiol 56:3835–3841
van Berkum P, Sloger C (1979) Immediate acetylene reduction by excised grass roots not previously preinoculated at low oxygen tensions. Plant Physiol 64:39–743
van Berkum P, Eardly BD (1998) Molecular Evolutionary Systematics of the Rhizobiaceae. In: Spaink H, Kondorosi A, Hooykaas P (eds) The Rhizobiaceae. Kluwer, Dordrecht, pp 1–24
van Berkum P, Fuhrmann JJ (2000) Evolutionary relationships among the soybean bradyrhizobia reconstructed from 16S rRNA gene and Internally Transcribed Spacer region sequence divergence. Int J Syst Evol Microbiol 50:2165–2172
van Berkum P, Navarro RB, Vargas AAT (1994) Classification of the uptake hydrogenase-positive (Hup+) bean rhizobia as Rhizobium tropici. Appl Environ Microbiol 60:554–561
van Berkum P, Beyene D, Eardly BD (1996) Phylogenetic relationships among Rhizobium species nodulating the common bean (Phaseolus vulgaris L.). Int J Syst Bacteriol 46:240–244
van Berkum P, Beyene D, Bao G, Campbell TA, Eardly BD (1998) Rhizobium mongolense sp. nov. is one of three rhizobial genotypes identified which nodulate and form nitrogen-fixing symbiosis with Medicago ruthenica [(L.) Ledebour]. Int J Syst Bacteriol 48:687–699
van Berkum P, Rehner S, Eardly BD (2001) How well does 16S rRNA gene phylogeny represent evolutionary relationships among the rhizobia? Proc of the 13th International Congress on Nitrogen Fixation, Hamilton, Canada
van Berkum P, Terefework Z, Paulin L, Suomalainen S, Lindstrom K, Eardly BD (2003) Discordant phylogenies within the rrn loci of rhizobia. J. Bacteriol 185:2988–2998
Vincent JM (1970) A manual for the practical study of root-nodule bacteria. International biological programme handbook. no. 15. Blackwell, Oxford
Willems A, Collins MD (1993) Phylogenetic analysis of rhizobia and agrobacteria based upon 16S rRNA gene sequences. Int J Syst Bacteriol 43:305–313
Young, JPW (1985) Rhizobium population genetics: enzyme polymorhism in isolates from peas, clover, beans and Lucerne grown in the same site. J Gen Microbiol 131:2399–2408
Acknowledgements
We especially thank the Institute of Agricultural Research for supplying the soil samples used in the study. We also thank K. Lee Nash and Patrick Elia for expert technical assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Beyene, D., Kassa, S., Ampy, F. et al. Ethiopian soils harbor natural populations of rhizobia that form symbioses with common bean (Phaseolus vulgaris L.). Arch Microbiol 181, 129–136 (2004). https://doi.org/10.1007/s00203-003-0636-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-003-0636-2