Abstract
Summary
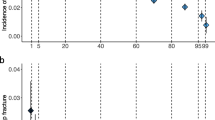
The potential of bone mineral density (BMD)-related genome-wide polygenic score (PGS) in identifying individuals with a high risk of fractures remains unclear. This study suggests that an efficient PGS enables the identification of strata with up to a 1.5-fold difference in fracture incidence. Incorporating PGS into clinical diagnosis is anticipated to increase the population-level screening benefits.
Purpose
This study sought to construct genome-wide polygenic scores for femoral neck and total body BMD and to estimate their potential in identifying individuals with a high risk of osteoporotic fractures.
Methods
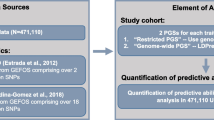
Genome-wide polygenic scores were developed and validated for femoral neck and total body BMD. We externally tested the PGSs, both by themselves and in combination with available clinical risk factors, in 455,663 European ancestry individuals from the UK Biobank. The predictive accuracy of the developed genome-wide PGS was also compared with previously published restricted PGS employed in fracture risk assessment.
Results
For each unit decrease in PGSs, the genome-wide PGSs were associated with up to 1.17-fold increased fracture risk. Out of four studied PGSs, \({\varvec{P}}{\varvec{G}}{\varvec{S}}\_{{\varvec{T}}{\varvec{B}}{\varvec{B}}{\varvec{M}}{\varvec{D}}}_{81}\) (HR: 1.03; 95%CI 1.01–1.05, p = 0.001) had the weakest and the \({\varvec{P}}{\varvec{G}}{\varvec{S}}\_{{\varvec{T}}{\varvec{B}}{\varvec{B}}{\varvec{M}}{\varvec{D}}}_{{\varvec{l}}{\varvec{d}}{\varvec{p}}{\varvec{r}}{\varvec{e}}{\varvec{d}}}\) (HR: 1.17; 95%CI 1.15–1.19, p < 0.0001) had the strongest association with an incident fracture. In the reclassification analysis, compared to the FRAX base model, the models with \({\varvec{P}}{\varvec{G}}{\varvec{S}}\_{{\varvec{F}}{\varvec{N}}{\varvec{B}}{\varvec{M}}{\varvec{D}}}_{63}\), \({\varvec{P}}{\varvec{G}}{\varvec{S}}\_{{\varvec{T}}{\varvec{B}}{\varvec{B}}{\varvec{M}}{\varvec{D}}}_{81}\), \({\varvec{P}}{\varvec{G}}{\varvec{S}}\_{{\varvec{F}}{\varvec{N}}{\varvec{B}}{\varvec{M}}{\varvec{D}}}_{{\varvec{l}}{\varvec{d}}{\varvec{p}}{\varvec{r}}{\varvec{e}}{\varvec{d}}}\), and \({\varvec{P}}{\varvec{G}}{\varvec{S}}\_{{\varvec{T}}{\varvec{B}}{\varvec{B}}{\varvec{M}}{\varvec{D}}}_{{\varvec{l}}{\varvec{d}}{\varvec{p}}{\varvec{r}}{\varvec{e}}{\varvec{d}}}\) improved the reclassification of fracture by 1.2% (95% CI, 1.0 to 1.3%), 0.2% (95% CI, 0.1 to 0.3%), 1.4% (95% CI, 1.3 to 1.5%), and 2.2% (95% CI, 2.1 to 2.4%), respectively.
Conclusions
Our findings suggested that an efficient PGS estimate enables the identification of strata with up to a 1.7-fold difference in fracture incidence. Incorporating PGS information into clinical diagnosis is anticipated to increase the benefits of screening programs at the population level.




Similar content being viewed by others
Data availability
Data sharing does not apply to this article as no datasets were generated during the current study.
References
Sözen T, Özışık L, Başaran NÇ (2017) An overview and management of osteoporosis. Eur J Rheumatol 4:46–56. https://doi.org/10.5152/eurjrheum.2016.048
Johnell O, Kanis JA (2006) An estimate of the worldwide prevalence and disability associated with osteoporotic fractures. Osteoporos Int 17:1726–1733. https://doi.org/10.1007/s00198-006-0172-4
Burge R, Dawson-Hughes B, Solomon DH, Wong JB, King A, Tosteson A (2007) Incidence and economic burden of osteoporosis-related fractures in the United States, 2005–2025. J Bone Miner Res 22:465–475. https://doi.org/10.1359/jbmr.061113
Kanis JA, Johnell O, Oden A, Johansson H, McCloskey E (2008) FRAX and the assessment of fracture probability in men and women from the UK. Osteoporos Int 19:385–397. https://doi.org/10.1007/s00198-007-0543-5
Briot K, Paternotte S, Kolta S, Eastell R, Felsenberg D, Reid DM, Glüer C, Roux C (2014) FRAX®: Prediction of major osteoporotic fractures in women from the general population: the OPUS study. PLoS One 8:e83436
Crandall CJ, Schousboe JT, Morin SN, Lix LM, Leslie W (2019) Performance of FRAX and FRAX-based treatment thresholds in women aged 40 years and older: the Manitoba BMD Registry. J Bone Miner Res 34:1419–1427. https://doi.org/10.1002/jbmr.3717
Sornay-Rendu E, Munoz F, Delmas PD, Chapurlat RD (2010) The FRAX tool in French women: how well does it describe the real incidence of fracture in the OFELY cohort? J Bone Miner Res 25:2101–2107. https://doi.org/10.1002/jbmr.106
Trajanoska K, Rivadeneira F (2019) The genetic architecture of osteoporosis and fracture risk. Bone 126:2–10
Ralston SH, Uitterlinden AG (2010) Genetics of osteoporosis. Endocr Rev 31:629–662. https://doi.org/10.1210/er.2009-0044
Wagner H, Melhus H, Pedersen NL, Michaëlsson K (2012) Heritable and environmental factors in the causation of clinical vertebral fractures. Calcif Tissue Int 90:458–464. https://doi.org/10.1007/s00223-012-9592-7
Andrew T, Antioniades L, Scurrah KJ, Macgregor AJ, Spector TD (2005) Risk of wrist fracture in women is heritable and is influenced by genes that are largely independent of those influencing BMD. J Bone Miner Res 20:67–74. https://doi.org/10.1359/JBMR.041015
Michaëlsson K, Melhus H, Ferm H, Ahlbom A, Pedersen NL (2005) Genetic liability to fractures in the elderly. Arch Intern Med 165:1825–1830
Arden NK, Spector TD (1997) Genetic influences on muscle strength, lean body mass, and bone mineral density: a twin study. J Bone Miner Res 12:2076–2081. https://doi.org/10.1359/jbmr.1997.12.12.2076
Medina-Gomez C, Kemp JP, Trajanoska K, Luan J, Chesi A, Ahluwalia TS, Mook-Kanamori DO, Ham A, Hartwig FP, Evans DS, Joro R, Nedeljkovic I, Zheng HF, Zhu K, Atalay M, Liu CT, Nethander M, Broer L, Porleifsson G, Mullin BH, Handelman SK, Nalls MA, Jessen LE, Heppe DHM, Richards JB, Wang C, Chawes B, Schraut KE, Amin N, Wareham N, Karasik D, Van der Velde N, Ikram MA, Zemel BS, Zhou Y, Carlsson CJ, Liu Y, McGuigan FE, Boer CG, Bønnelykke K, Ralston SH, Robbins JA, Walsh JP, Zillikens MC, Langenberg C, Li-Gao R, Williams FMK, Harris TB, Akesson K, Jackson RD, Sigurdsson G, den Heijer M, van der Eerden BCJ, van de Peppel J, Spector TD, Pennell C, Horta BL, Felix JF, Zhao JH, Wilson SG, de Mutsert R, Bisgaard H, Styrkársdóttir U, Jaddoe VW, Orwoll E, Lakka TA, Scott R, Grant SFA, Lorentzon M, van Duijn CM, Wilson JF, Stefansson K, Psaty BM, Kiel DP, Ohlsson C, Ntzani E, van Wijnen AJ, Forgetta V, Ghanbari M, Logan JG, Williams GR, Bassett JHD, Croucher PI, Evangelou E, Uitterlinden AG, Ackert-Bicknell CL, Tobias JH, Evans DM, Rivadeneira F (2018) Life-course genome-wide association study meta-analysis of total body BMD and assessment of age-specific effects. Am J Hum Genet 102:88–102
Kim SK (2018) Identification of 613 new loci associated with heel bone mineral density and a polygenic risk score for bone mineral density, osteoporosis and fracture. PLoS One 13:e0200785
Estrada K, Styrkarsdottir U, Evangelou E, Hsu Y, Duncan EL, Ntzani EE, Oei L, Albagha OME, Amin N, Kemp JP, Koller DL, Li G, Liu C, Minster RL, Moayyeri A, Vandenput L, Willner D, Xiao S, Yerges-Armstrong L, Zheng H, Alonso N, Eriksson J, Kammerer CM, Kaptoge SK, Leo PJ, Thorleifsson G, Wilson SG, Wilson JF, Aalto V, Alen M, Aragaki AK, Aspelund T, Center JR, Dailiana Z, Duggan DJ, Garcia M, Garcia-Giralt N, Giroux S, Hallmans G, Hocking LJ, Husted LB, Jameson KA, Khusainova R, Kim GS, Kooperberg C, Koromila T, Kruk M, Laaksonen M, Lacroix AZ, Lee SH, Leung PC, Lewis JR, Masi L, Mencej-Bedrac S, Nguyen TV, Nogues X, Patel MS, Prezelj J, Rose LM, Scollen S, Siggeirsdottir K, Smith AV, Svensson O, Trompet S, Trummer O, van Schoor NM, Woo J, Zhu K, Balcells S, Brandi ML, Buckley BM, Cheng S, Christiansen C, Cooper C, Dedoussis G, Ford I, Frost M, Goltzman D, González-Macías J, Kähönen M, Karlsson M, Khusnutdinova E, Koh J, Kollia P, Langdahl BL, Leslie WD, Lips P, Ljunggren Ö, Lorenc RS, Marc J, Mellström D, Obermayer-Pietsch B, Olmos JM, Pettersson-Kymmer U, Reid DM, Riancho JA, Ridker PM, Rousseau F, Slagboom PE, Tang NLS, Urreizti R, Van Hul W, Viikari J, Zarrabeitia MT, Aulchenko YS, Castano-Betancourt M, Grundberg E, Herrera L, Ingvarsson T, Johannsdottir H, Kwan T, Li R, Luben R, Medina-Gómez C, Palsson ST, Reppe S, Rotter JI, Sigurdsson G, van Meurs JBJ, Verlaan D, Williams FMK, Wood AR, Zhou Y, Gautvik KM, Pastinen T, Raychaudhuri S, Cauley JA, Chasman DI, Clark GR, Cummings SR, Danoy P, Dennison EM, Eastell R, Eisman JA, Gudnason V, Hofman A, Jackson RD, Jones G, Jukema JW, Khaw K, Lehtimäki T, Liu Y, Lorentzon M, McCloskey E, Mitchell BD, Nandakumar K, Nicholson GC, Oostra BA, Peacock M, Pols HAP, Prince RL, Raitakari O, Reid IR, Robbins J, Sambrook PN, Sham PC, Shuldiner AR, Tylavsky FA, van Duijn CM, Wareham NJ, Cupples LA, Econs MJ, Evans DM, Harris TB, Kung AWC, Psaty BM, Reeve J, Spector TD, Streeten EA, Zillikens MC, Thorsteinsdottir U, Ohlsson C, Karasik D, Richards JB, Brown MA, Stefansson K, Uitterlinden AG, Ralston SH, Ioannidis JPA, Kiel DP, Rivadeneira F (2012) Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat Genet 44:491–501. https://doi.org/10.1038/ng.2249
Morris JA, Kemp JP, Youlten SE, Laurent L, Logan JG, Chai RC, Vulpescu NA, Forgetta V, Kleinman A, Mohanty ST, Sergio CM, Quinn J, Nguyen-Yamamoto L, Luco A, Vijay J, Simon M, Pramatarova A, Medina-Gomez C, Trajanoska K, Ghirardello EJ, Butterfield NC, Curry KF, Leitch VD, Sparkes PC, Adoum A, Mannan NS, Komla-Ebri D, Pollard AS, Dewhurst HF, Hassall TAD, Beltejar MG, 23andMe RT, Adams DJ, Vaillancourt SM, Kaptoge S, Baldock P, Cooper C, Reeve J, Ntzani EE, Evangelou E, Ohlsson C, Karasik D, Rivadeneira F, Kiel DP, Tobias JH, Gregson CL, Harvey NC, Grundberg E, Goltzman D, Adams DJ, Lelliott CJ, Hinds DA, Ackert-Bicknell C, Hsu Y, Maurano MT, Croucher PI, Williams GR, Bassett JHD, Evans DM, Richards JB (2019) An atlas of genetic influences on osteoporosis in humans and mice. Nat Genet 51:258–66. https://doi.org/10.1038/s41588-018-0302-x
Lewis CM, Vassos E (2020) Polygenic risk scores: from research tools to clinical instruments. Genome Med 12:44. https://doi.org/10.1186/s13073-020-00742-5
Eriksson J, Evans DS, Nielson CM, Shen J, Srikanth P, Hochberg M, McWeeney S, Cawthon PM, Wilmot B, Zmuda J, Tranah G, Mirel DB, Challa S, Mooney M, Crenshaw A, Karlsson M, Mellström D, Vandenput L, Orwoll E, Ohlsson C (2015) Limited clinical utility of a genetic risk score for the prediction of fracture risk in elderly subjects. J Bone Miner Res 30:184–194. https://doi.org/10.1002/jbmr.2314
Warrington NM, Kemp JP, Tilling K, Tobias JH, Evans DM (2015) Genetic variants in adult bone mineral density and fracture risk genes are associated with the rate of bone mineral density acquisition in adolescence. Hum Mol Genet 24:4158–4166. https://doi.org/10.1093/hmg/ddv143
Nethander M, Pettersson-Kymmer U, Vandenput L, Lorentzon M, Karlsson M, Mellström D, Ohlsson C (2020) BMD-related genetic risk scores predict site-specific fractures as well as trabecular and cortical bone microstructure. J Clin Endocrinol Metab 105:1344
Xiao X, Wu Q (2021) The utility of genetic risk score to improve performance of FRAX for fracture prediction in US postmenopausal women. Calcif Tissue Int 108:746–756. https://doi.org/10.1007/s00223-021-00809-4
Ho-Le TP, Center JR, Eisman JA, Nguyen HT, Nguyen TV (2017) Prediction of bone mineral density and fragility fracture by genetic profiling. J Bone Miner Res 32:285–293. https://doi.org/10.1002/jbmr.2998
Mitchell JA, Chesi A, Elci O, McCormack SE, Roy SM, Kalkwarf HJ, Lappe JM, Gilsanz V, Oberfield SE, Shepherd JA, Kelly A, Grant SF, Zemel BS (2016) Genetic risk scores implicated in adult bone fragility associate with pediatric bone density. J Bone Miner Res 31:789–795. https://doi.org/10.1002/jbmr.2744
Mavaddat N, Michailidou K, Dennis J, Lush M, Fachal L, Lee A, Tyrer JP, Chen TH, Wang Q, Bolla MK, Yang X, Adank MA, Ahearn T, Aittomäki K, Allen J, Andrulis IL, Anton-Culver H, Antonenkova NN, Arndt V, Aronson KJ, Auer PL, Auvinen P, Barrdahl M, Beane Freeman LE, Beckmann MW, Behrens S, Benitez J, Bermisheva M, Bernstein L, Blomqvist C, Bogdanova NV, Bojesen SE, Bonanni B, Børresen-Dale AL, Brauch H, Bremer M, Brenner H, Brentnall A, Brock IW, Brooks-Wilson A, Brucker SY, Brüning T, Burwinkel B, Campa D, Carter BD, Castelao JE, Chanock SJ, Chlebowski R, Christiansen H, Clarke CL, Collée JM, Cordina-Duverger E, Cornelissen S, Couch FJ, Cox A, Cross SS, Czene K, Daly MB, Devilee P, Dörk T, Dos-Santos-Silva I, Dumont M, Durcan L, Dwek M, Eccles DM, Ekici AB, Eliassen AH, Ellberg C, Engel C, Eriksson M, Evans DG, Fasching PA, Figueroa J, Fletcher O, Flyger H, Försti A, Fritschi L, Gabrielson M, Gago-Dominguez M, Gapstur SM, García-Sáenz JA, Gaudet MM, Georgoulias V, Giles GG, Gilyazova IR, Glendon G, Goldberg MS, Goldgar DE, González-Neira A, Grenaker Alnæs GI, Grip M, Gronwald J, Grundy A, Guénel P, Haeberle L, Hahnen E, Haiman CA, Håkansson N, Hamann U, Hankinson SE, Harkness EF, Hart SN, He W, Hein A, Heyworth J, Hillemanns P, Hollestelle A, Hooning MJ, Hoover RN, Hopper JL, Howell A, Huang G, Humphreys K, Hunter DJ, Jakimovska M, Jakubowska A, Janni W, John EM, Johnson N, Jones ME, Jukkola-Vuorinen A, Jung A, Kaaks R, Kaczmarek K, Kataja V, Keeman R, Kerin MJ, Khusnutdinova E, Kiiski JI, Knight JA, Ko YD, Kosma VM, Koutros S, Kristensen VN, Krüger U, Kühl T, Lambrechts D, Le Marchand L, Lee E, Lejbkowicz F, Lilyquist J, Lindblom A, Lindström S, Lissowska J, Lo WY, Loibl S, Long J, Lubiński J, Lux MP, MacInnis RJ, Maishman T, Makalic E, Maleva Kostovska I, Mannermaa A, Manoukian S, Margolin S, Martens JWM, Martinez ME, Mavroudis D, McLean C, Meindl A, Menon U, Middha P, Miller N, Moreno F, Mulligan AM, Mulot C, Muñoz-Garzon VM, Neuhausen SL, Nevanlinna H, Neven P, Newman WG, Nielsen SF, Nordestgaard BG, Norman A, Offit K, Olson JE, Olsson H, Orr N, Pankratz VS, Park-Simon TW, Perez JIA, Pérez-Barrios C, Peterlongo P, Peto J, Pinchev M, Plaseska-Karanfilska D, Polley EC, Prentice R, Presneau N, Prokofyeva D, Purrington K, Pylkäs K, Rack B, Radice P, Rau-Murthy R, Rennert G, Rennert HS, Rhenius V, Robson M, Romero A, Ruddy KJ, Ruebner M, Saloustros E, Sandler DP, Sawyer EJ, Schmidt DF, Schmutzler RK, Schneeweiss A, Schoemaker MJ, Schumacher F, Schürmann P, Schwentner L, Scott C, Scott RJ, Seynaeve C, Shah M, Sherman ME, Shrubsole MJ, Shu XO, Slager S, Smeets A, Sohn C, Soucy P, Southey MC, Spinelli JJ, Stegmaier C, Stone J, Swerdlow AJ, Tamimi RM, Tapper WJ, Taylor JA, Terry MB, Thöne K, Tollenaar RAEM, Tomlinson I, Truong T, Tzardi M, Ulmer HU, Untch M, Vachon CM, van Veen EM, Vijai J, Weinberg CR, Wendt C, Whittemore AS, Wildiers H, Willett W, Winqvist R, Wolk A, Yang XR, Yannoukakos D, Zhang Y, Zheng W, Ziogas A, ABCTB Investigators, kConFab/AOCS Investigators, NBCS Collaborators, Dunning AM, Thompson DJ, Chenevix-Trench G, Chang-Claude J, Schmidt MK, Hall P, Milne RL, Pharoah PDP, Antoniou AC, Chatterjee N, Kraft P, García-Closas M, Simard J, Easton DF (2019) Polygenic risk scores for prediction of breast cancer and breast cancer subtypes. Am J Hum Genet 104:21–34
Agerbo E, Sullivan PF, Vilhjálmsson BJ, Pedersen CB, Mors O, Børglum AD, Hougaard DM, Hollegaard MV, Meier S, Mattheisen M, Ripke S, Wray NR, Mortensen PB (2015) Polygenic risk score, parental socioeconomic status, family history of psychiatric disorders, and the risk for schizophrenia: a Danish population-based study and meta-analysis. JAMA Psychiat 72:635–641. https://doi.org/10.1001/jamapsychiatry.2015.0346
Inouye M, Abraham G, Nelson CP, Wood AM, Sweeting MJ, Dudbridge F, Lai FY, Kaptoge S, Brozynska M, Wang T, Ye S, Webb TR, Rutter MK, Tzoulaki I, Patel RS, Loos RJF, Keavney B, Hemingway H, Thompson J, Watkins H, Deloukas P, Di Angelantonio E, Butterworth AS, Danesh J, Samani NJ (2018) Genomic risk prediction of coronary artery disease in 480,000 adults: implications for primary prevention. J Am Coll Cardiol 72:1883–1893. https://doi.org/10.1016/j.jacc.2018.07.079
Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, Downey P, Elliott P, Green J, Landray M, Liu B, Matthews P, Ong G, Pell J, Silman A, Young A, Sprosen T, Peakman T, Collins R (2015) UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med 12:e1001779
Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, Motyer A, Vukcevic D, Delaneau O, O’Connell J, Cortes A, Welsh S, Young A, Effingham M, McVean G, Leslie S, Allen N, Donnelly P, Marchini J (2018) The UK Biobank resource with deep phenotyping and genomic data. Nature 562:203–209. https://doi.org/10.1038/s41586-018-0579-z
McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, Kang HM, Fuchsberger C, Danecek P, Sharp K, Luo Y, Sidore C, Kwong A, Timpson N, Koskinen S, Vrieze S, Scott LJ, Zhang H, Mahajan A, Veldink J, Peters U, Pato C, van Duijn CM, Gillies CE, Gandin I, Mezzavilla M, Gilly A, Cocca M, Traglia M, Angius A, Barrett JC, Boomsma D, Branham K, Breen G, Brummett CM, Busonero F, Campbell H, Chan A, Chen S, Chew E, Collins FS, Corbin LJ, Smith GD, Dedoussis G, Dorr M, Farmaki AE, Ferrucci L, Forer L, Fraser RM, Gabriel S, Levy S, Groop L, Harrison T, Hattersley A, Holmen OL, Hveem K, Kretzler M, Lee JC, McGue M, Meitinger T, Melzer D, Min JL, Mohlke KL, Vincent JB, Nauck M, Nickerson D, Palotie A, Pato M, Pirastu N, McInnis M, Richards JB, Sala C, Salomaa V, Schlessinger D, Schoenherr S, Slagboom PE, Small K, Spector T, Stambolian D, Tuke M, Tuomilehto J, Van den Berg LH, Van Rheenen W, Volker U, Wijmenga C, Toniolo D, Zeggini E, Gasparini P, Sampson MG, Wilson JF, Frayling T, de Bakker PI, Swertz MA, McCarroll S, Kooperberg C, Dekker A, Altshuler D, Willer C, Iacono W, Ripatti S, Soranzo N, Walter K, Swaroop A, Cucca F, Anderson CA, Myers RM, Boehnke M, McCarthy MI, Durbin R, Haplotype Reference Consortium (2016) A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet 48:1279–83. https://doi.org/10.1038/ng.3643
Fry A, Littlejohns TJ, Sudlow C, Doherty N, Adamska L, Sprosen T, Collins R, Allen NE (2017) Comparison of sociodemographic and health-related characteristics of UK Biobank participants with those of the general population. Am J Epidemiol 186:1026–1034. https://doi.org/10.1093/aje/kwx246
Privé F, Arbel J, Vilhjálmsson BJ (2020) LDpred2: better, faster, stronger. Bioinformatics 36:5424–5431. https://doi.org/10.1093/bioinformatics/btaa1029
Schoenfeld D (1982) Partial residuals for the proportional hazards regression model. Biometrika 69:239–241. https://doi.org/10.1093/biomet/69.1.239
Therneau TM, Grambsch PM, Fleming TR (1990) Martingale-based residuals for survival models. Biometrika 77:147–160. https://doi.org/10.1093/biomet/77.1.147
Zheng H, Forgetta V, Hsu Y, Estrada K, Rosello-Diez A, Leo PJ, Dahia CL, Park-Min KH, Tobias JH, Kooperberg C, Kleinman A, Styrkarsdottir U, Liu C, Uggla C, Evans DS, Nielson CM, Walter K, Pettersson-Kymmer U, McCarthy S, Eriksson J, Kwan T, Jhamai M, Trajanoska K, Memari Y, Min J, Huang J, Danecek P, Wilmot B, Li R, Chou W, Mokry LE, Moayyeri A, Claussnitzer M, Cheng C, Cheung W, Medina-Gómez C, Ge B, Chen S, Choi K, Oei L, Fraser J, Kraaij R, Hibbs MA, Gregson CL, Paquette D, Hofman A, Wibom C, Tranah GJ, Marshall M, Gardiner BB, Cremin K, Auer P, Hsu L, Ring S, Tung JY, Thorleifsson G, Enneman AW, van Schoor NM, de Groot, Lisette C P G M, van der Velde N, Melin B, Kemp JP, Christiansen C, Sayers A, Zhou Y, Calderari S, van Rooij J, Carlson C, Peters U, Berlivet S, Dostie J, Uitterlinden AG, Williams SR, Farber C, Grinberg D, LaCroix AZ, Haessler J, Chasman DI, Giulianini F, Rose LM, Ridker PM, Eisman JA, Nguyen TV, Center JR, Nogues X, Garcia-Giralt N, Launer LL, Gudnason V, Mellström D, Vandenput L, Amin N, van Duijn CM, Karlsson MK, Ljunggren Ö, Svensson O, Hallmans G, Rousseau F, Giroux S, Bussière J, Arp PP, Koromani F, Prince RL, Lewis JR, Langdahl BL, Hermann AP, Jensen JB, Kaptoge S, Khaw K, Reeve J, Formosa MM, Xuereb-Anastasi A, Åkesson K, McGuigan FE, Garg G, Olmos JM, Zarrabeitia MT, Riancho JA, Ralston SH, Alonso N, Jiang X, Goltzman D, Pastinen T, Grundberg E, Gauguier D, Orwoll ES, Karasik D, Davey-Smith G, AOGC Consortium, Smith AV, Siggeirsdottir K, Harris TB, Zillikens MC, van Meurs JBJ, Thorsteinsdottir U, Maurano MT, Timpson NJ, Soranzo N, Durbin R, Wilson SG, Ntzani EE, Brown MA, Stefansson K, Hinds DA, Spector T, Cupples LA, Ohlsson C, Greenwood CMT, UK10K Consortium, Jackson RD, Rowe DW, Loomis CA, Evans DM, Ackert-Bicknell CL, Joyner AL, Duncan EL, Kiel DP, Rivadeneira F, Richards JB (2015) Whole-genome sequencing identifies EN1 as a determinant of bone density and fracture. Nature 526:112–7. https://doi.org/10.1038/nature14878
Kanis JA (1994) Assessment of fracture risk and its application to screening for postmenopausal osteoporosis: synopsis of a WHO report. WHO Study Group Osteoporos Int 4:368–381. https://doi.org/10.1007/BF01622200
Speed D, Cai N, UCLEB Consortium, Johnson MR, Nejentsev S, Balding DJ (2017) Reevaluation of SNP heritability in complex human traits. Nat Genet 49:986–992. https://doi.org/10.1038/ng.3865
Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, Madden PA, Heath AC, Martin NG, Montgomery GW, Goddard ME, Visscher PM (2010) Common SNPs explain a large proportion of the heritability for human height. Nat Genet 42:565–569. https://doi.org/10.1038/ng.608
Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, Natarajan P, Lander ES, Lubitz SA, Ellinor PT, Kathiresan S (2018) Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet 50:1219–1224. https://doi.org/10.1038/s41588-018-0183-z
Fritsche LG, Beesley LJ, VandeHaar P, Peng RB, Salvatore M, Zawistowski M, Gagliano Taliun SA, Das S, LeFaive J, Kaleba EO, Klumpner TT, Moser SE, Blanc VM, Brummett CM, Kheterpal S, Abecasis GR, Gruber SB, Mukherjee B (2019) Exploring various polygenic risk scores for skin cancer in the phenomes of the Michigan genomics initiative and the UK Biobank with a visual catalog: PRSWeb. PLoS Genet 15:e1008202. https://doi.org/10.1371/journal.pgen.1008202
Allegrini AG, Selzam S, Rimfeld K, von Stumm S, Pingault JB, Plomin R (2019) Genomic prediction of cognitive traits in childhood and adolescence. Mol Psychiatry 24:819–827. https://doi.org/10.1038/s41380-019-0394-4
Chung W, Chen J, Turman C, Lindstrom S, Zhu Z, Loh P, Kraft P, Liang L (2019) Efficient cross-trait penalized regression increases prediction accuracy in large cohorts using secondary phenotypes. Nat Commun 10:569. https://doi.org/10.1038/s41467-019-08535-0
Janssens ACJW, Joyner MJ (2019) Polygenic risk scores that predict common diseases using millions of single nucleotide polymorphisms: is more, better? Clin Chem 65:609–611. https://doi.org/10.1373/clinchem.2018.296103
Lee SH, Lee SW, Ahn SH, Kim T, Lim KH, Kim BJ, Cho EH, Kim SW, Kim TH, Kim GS, Kim SY, Koh JM, Kang C (2013) Multiple gene polymorphisms can improve prediction of nonvertebral fracture in postmenopausal women. J Bone Miner Res 28:2156–2164. https://doi.org/10.1002/jbmr.1955
Lu T, Forgetta V, Keller-Baruch J, Nethander M, Bennett D, Forest M, Bhatnagar S, Walters RG, Lin K, Chen Z (2021) Improved prediction of fracture risk leveraging a genome-wide polygenic risk score. Genome medicine 13:1–15
Acknowledgements
The National Supercomputing Institute at the University of Nevada Las Vegas provided facilities for bioinformatical analysis in this study. Part of Dr. Qing Wu’s work was completed at the Nevada Institute of Personalized Medicine, College of Sciences and at the Department of Epidemiology and Biostatistics, School of Public Health, University of Nevada, Las Vegas.
Funding
The study was supported by the National Institute of General Medical Sciences under award number P20GM121325 and by the National Institute on Minority Health and Health Disparities of the National Institutes of Health under award number 1R21MD013681.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics approval
This research work was approved by the UK Biobank and the institutional review board at the University of Nevada, Las Vegas. This study has been conducted using the UK Biobank Data Resource under application number 58122.
Conflict of interest
None.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xiao, X., Wu, Q. The clinical utility of the BMD-related comprehensive genome-wide polygenic score in identifying individuals with a high risk of osteoporotic fractures. Osteoporos Int 34, 681–692 (2023). https://doi.org/10.1007/s00198-022-06654-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00198-022-06654-x