Abstract
Introduction and hypothesis
Growing literature details the critical importance of the microbiome in the modulation of human health and disease including both the gastrointestinal and genitourinary systems. Rectovaginal fistulae (RVF) are notoriously difficult to manage, many requiring multiple attempts at repair before correction is achieved. RVF involves two distinct microbiome communities whose characteristics and potential interplay have not been previously characterized and may influence surgical success.
Methods
In this pilot study, rectal and vaginal samples were collected from 14 patients with RVF. Samples were collected preoperatively, immediately following surgery, 6–8 weeks postoperatively and at the time of any fistula recurrence. Amplification of the 16S rDNA V3-V5 gene region was done to identify microbiota. Data were summarized using both α-diversity to describe species richness and evenness and β-diversity to characterize the shared variation between communities. Differential abundance analysis was performed to identify microbial taxa associated with recurrence.
Results
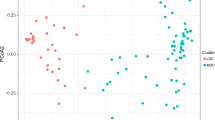
The rectal and vaginal microbiome in patients undergoing successful fistula repair was different than in those with recurrence (β-diversity, p = 0.005 and 0.018, respectively) and was characterized by higher species diversity (α-diversity, p = 0.07 and p = 0.006, respectively). Thirty-one taxa were enriched in patients undergoing successful repair to include Bacteroidetes, Alistipes and Rikenellaceae as well as Firmicutes, Subdoligranulum, Ruminococcaceae UCG-010 and NK4A214 group.
Conclusions
Microbiome characteristics associated with fistula recurrence have been identified. The association of higher vaginal diversity with a favorable outcome has not been previously described. Expansion of this pilot project is needed to confirm findings. Taxa associated with successful repair could be targeted for subsequent therapeutic intervention.




Similar content being viewed by others
References
Pinto RA, Peterson TV, Shawki S, Davila GW, Wexner SD. Are there predictors of outcome following rectovaginal fistula repair? Dis Colon Rectum. 2010;53(9):1240–7.
deSouza A, Abcarian H. The Management of Rectovaginal Fistula. In: Cameron JL, Cameron AM, editors. Current surgical therapy. 11th ed. Philadelphia: Saunders; 2014. p. 283–8.
United Nations Population Fund (2015) Obstetric Fistula 2015, from http://www.unfpa.org/obstetric-fistula. Accessed Nov 2017
Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449(7164):804–10.
Wexner SD, Ruiz DE, Genua J, et al. Gracilis muscle interposition for the treatment of rectourethral, rectovaginal, and pouch-vaginal fistulas; results in 53 patients. Ann Surg. 2008;248(1):39–43.
Das B, Snyder M. Rectovaginal fistulae. Clin Colon Rectal Surg. 2016;29:50–6.
Jones RS, Stukenborg GJ. Patient-reported outcomes measurement information system (PROMIS) use in surgical care: a scoping study. J Am Coll Surg. 2017;224(3):245–54.
Callahan BJ, McMurdie PJ, Rosen MJ, et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13(7):581.
Quast C, Pruesse E, Yilmaz P, Gerken J, Glöckner FO. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2012;41(D1):D590–6.
Price MN, Dehal PS, Arkin AP. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5(3):9490.
Chen J, Bittinger K, Charlson ES, et al. Associating microbiome composition with environmental covariates using generalized UniFrac distances. Bioinformatics. 2012;28(16):2106–13.
Chen L, Reeve J, Zhang L, et al. GMPR: a robust normalization method for zero-inflated count data with application to microbiome sequencing data. PeerJ. 2018;6:e4600.
Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–14.
Ma B, Forney LL, Ravel J. The vaginal microbiome: rethinking health and diseases. Annu Rev Microbiol. 2012;66:371–89.
Hyman RW, Fukushima M, Diamond L, Kumm J, Guidice LC, Davis RW. Microbes of the human vaginal epithelium. Proc Natl Acad Sci U S A. 2005;102(22):7952–7.
Kim TK, Thomas SM, Ho M, et al. Heterogeneity of vaginal microbial communities within individuals. J Clin Microbiol. 2009;47(4):1181–9.
Ravel J, Pawel G, Abdo Z, et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A. 2011;108(S1):4680–7.
White BA, Creedon DJ, Nelson KE, Wilson BA. The vaginal microbiome in health and disease. Trends Endocrinol Metab. 2011;22(10):389–93.
Bayigga L, Kateete DP, Anderson DJ, et al. Diversity of vaginal microbiota in sub-Saharan Africa and its effects on HIV transmission and prevention. AJOG. 2019;220(2):155–66.
Bordgorff H, Tsivtsivadze E, Verhelst R, et al. Lactobacillus-dominated cervicovaginal microbiota associated with reduced HIV/STI prevalence and genital HIV viral load in African women. ISME J. 2014;8:1781–93.
Wang H, Ma Y, Li R, et al. Associations of Cervicovaginal lactobacilli with high-risk human papillomavirus infection, cervical intraepithelial neoplasia, and cancer: a systematic review and meta-analysis. JID. 2019;220:1243–54.
Dols JA, Smit PW, Kort R, et al. PCR-based identification of eight Lactobacillus species and 18 HR-HPV genotypes in fixed cervical samples of south African women at risk of HIV and BV. Diagn Cytopathol. 40:472–7.
Spear GT, Sikaroodi M, Zariffard MR, et al. Copmarison of the diversity of the vaginal microbiota in HIV-infected and HIV-uninfected women with or without bacterial vaginosis. J Infect Dis. 198:1131–40.
Mitchell C, Balkus JE, Fredricks D, et al. Interaction between lactobacilli, bacterial vaginosis-associated bacteria, and HIV type 1 RNA and DNA genital shedding in US and Kenyan women. AIDS Res Hum Retrovir. 29:13–9.
Chang JY, Antonopoulos DA, Kalra A, Tonelli A, Khalife WT, Schmidt TM, et al. Decreased diversity of the fecal microbiome in recurrent Clostridium dificile-associated diarrhea. J Infect Dis. 2008;197(3):435–8.
Ott SJ, Musfeldt M, Wenderoth DF, et al. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut. 2004;53(5):685–93.
Gong D, Gong X, Wang L, Yu X, Dong Q. Involvement of reduced microbial diversity in inflammatory bowel disease. Gastroenterol Res Pract 2016;2016:6951091.
Verhoog S, Taneri PE Diaz ZM, et al. Dietary factors and modulation of Bacteria strains of Akkermansia muciniphila and Faecalibacterium prausnitzii; a systematic review. Nutrients. 2019;11(7). https://doi.org/10.3390/nu11071565,10.3390/nu11071565.
Montassier E, Al-Ghalith AG, Ward T, et al. Pretreatment gut microbiome predicts chemotherapy-related bloodstream infection. Genome Med. 2016;8:49.
Dinh DM, Volpe GE, Duffalo C, et al. Intestinal microbiota, microbial translocation, and systemic inflammation in chronic HIV infection. J Infect Dis. 2015;211(1):19–27.
Dubin K, Callahan MK, Ren B, et al. Intestinal microbioime analysis identify melanoma patients at risk for checkpoint-blockade-induced colitis. Nat Commun. 2016;7:10391.
Chase D, Goulder A, Zenhausern F, et al. The vaginal and gastrointestinal microbiomes in gynecologic cancers: a review of applications in etiology, symptoms and treatment. Gynecol Oncol. 2015;138(1):190–200.
Lee P, Br Y, Yacyshyn MB. Gut microbiota and obesity; an opportunity to alter obesity through faecal microbiota transplant (FMT). Diabetes Obes Metab. 2019;21(3):479–49.
Russel SL Gold MJ, Hartmann M, et al. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. 2012;13(5):440–7.
Nylund L, Nermes M, Isolauri E, et al. Severity of atopic disease inversely correlates with intestinal microbiota diversity and butyrate-producing bacteria. Allergy. 2015;70(2):241–4.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
This work was supported by a grant from the International Urogynecological Association.
This project was supported by CTSA grant number KL2 TR002379 from the National Center for Advancing Translational Science (NCATS) and was also supported, in part, by a career enhancement award from NIH grant P50 CA136393. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
MWA is a member of the scientific advisory board of LUCA Biologics, Inc., on research related to urinary tract infections, preterm birth and reproductive medicine. These activities do not overlap with the research presented here.
Dr. Leach is a member of the US Armed Forces; the views and opinions expressed in this article are those of the authors and do not necessarily reflect the official policy or position of this government agency.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementay Information
ESM 1
(DOCX 1222 kb)
Rights and permissions
About this article
Cite this article
Leach, D.A., Chen, J., Yang, L. et al. Microbiome diversity predicts surgical success in patients with rectovaginal fistula. Int Urogynecol J 32, 2491–2501 (2021). https://doi.org/10.1007/s00192-020-04580-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00192-020-04580-2