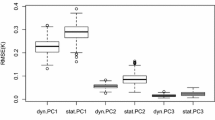
Abstract
Exploratory analysis and visualization of multiple time series data are essential for discovering the underlying dynamics of a series before attempting modeling and forecasting. This study extends two dimension reduction methods - principal component analysis (PCA) and sliced inverse regression (SIR) - to multiple time series data. This is achieved through the innovative path point approach, a new addition to the symbolic data analysis framework. By transforming multiple time series data into time-dependent intervals marked by starting and ending values, each series is geometrically represented as successive directed segments with unique path points. These path points serve as the foundation of our novel representation approach. PCA and SIR are then applied to the data table formed by the coordinates of these path points, enabling visualization of temporal trajectories of objects within a reduced-dimensional subspace. Empirical studies encompassing simulations, microarray time series data from a yeast cell cycle, and financial data confirm the effectiveness of our path point approach in revealing the structure and behavior of objects within a 2D factorial plane. Comparative analyses with existing methods, such as the applied vector approach for PCA and SIR on time-dependent interval data, further underscore the strength and versatility of our path point representation in the realm of time series data.










Similar content being viewed by others
References
Aigner W, Miksch S, Müller W, Schumann H, Tominski C (2007) Visualizing time-oriented data—a systematic view. Comput Graph 31(3):401–409
Bar-Joseph Z, Gitter A, Simon I (2012) Studying and modelling dynamic biological processes using time-series gene expression data. Nat Rev Genet 13(8):552–564
Becker C, Fried R (2003) Sliced inverse regression for high-dimensional time series. In: Exploratory data analysis in empirical research: proceedings of the 25th annual conference of the gesellschaft fur klassifickation, University of Munich. pp 3 – 11
Bertrand P, Goupil F (2000) Descriptive statistics for symbolic data. In: Bock HH, Diday E (eds) Analysis of symbolic data: exploratory methods for extracting statistical information from complex data. Springer-Verlag, Berlin, pp 103–124
Billard L (2008) Sample covariance functions for complex quantitative data. In: Mizuta M. and Nakano J. (Ed): Proceedings of the international association of statistical computing conference, pp 157 – 163. Yokohama
Billard L, Diday E (2003) From the statistics of data to the statistics of knowledge: symbolic data analysis. J Am Stat Assoc 98(462):470–487
Billard L, Diday E (2006) Symbolic data analysis: conceptual statistics and data mining. Wiley
Bock HH, Diday E (2000) Analysis of symbolic data: explanatory methods for extracting statistical information from complex data. Springer-Verlag, Berlin
Cazes P, Chouakria A, Diday E, Schecktman Y (1997) Extension de l’analyse en composantes principales ’a des donn’ees de type intervalle. Rev Stat Appl 45:5–24
Chen CH, Li KC (1998) Can SIR be as popular as multiple linear regression? Stat Sinica 8:289–316
Cho RJ et al (1998) A genomewide transcriptional analysis of the mitotic cell cycle. Mol Cell 2(1):65–73
Chouakria A (1998) Extension de l’analyse en composantes principales ’a des donn’ees de type intervalle. Doctoral thesis; University of Paris IX Dauphine
Cook RD (1994) On the interpretation of regression plots. J Am Stat Assoc 89:177–190
Cook RD (1996) Graphics for regressions with a binary response. J Am Stat Assoc 91:983–992
Cook RD (2000) SAVE: a method for dimension reduction and graphics in regression. Commun Stat Theor Methods 29:2109–2121
Cook RD, Critchley F (2000) Identifying regression outliers and mixtures graphically. J Am Stat Assoc 95:781–794
Cox TF, Cox MAA (2001) Multidimensional scaling. Chapman and Hall, London
Diday E (2016) Thinking by classes in data science: the symbolic data analysis paradigm. WIREs Comput Stat 8:172–205
Douzal-Chouakria A, Billard L, Diday E (2011) Principal component analysis for interval-valued observations. Stat Anal Data Min 4:229–246
D’Urso P, Giordani P (2004) A least squares approach to principal component analysis for interval valued data. Chem Intell Lab Syst 70:179–192
Ernst J, Nau GJ, Bar-Joseph Z (2005) Clustering short time series gene expression data. Bioinformatics 21(Suppl 1):i159-68
Fernández A, García S, Galar M, Prati RC, Krawczyk B, Herrera F (2018) Dimensionality reduction for imbalanced learning. In: Learning from imbalanced data sets. Springer, Cham
Ferre L (1998) Determining the dimension in sliced inverse regression and related methods. J Am Stat Assoc 93(441):132–140
Gioia F, Lauro NC (2006) Principal component analysis on interval data. Comput Stat 21:343–363
Giordani P, Kiers HAL (2006) A comparison of three methods for principal component analysis of fuzzy interval data. Comput Stati Data Anal 51:379–397
Gracia A, Gonzalez S, Robles V, Menasalvas E (2014) A methodology to compare dimensionality reduction algorithms in terms of loss of quality. Inf Sci 270:1–27
Ichino M (2011) The quantile method for symbolic principal component analysis. Stat Anal Data Min 4(2):184–198
Irpino A (2006) Spaghetti PCA analysis: an extension of principal components analysis to time dependent interval data. Pattern Recogn Lett 27:504–513
Irpino A (2013) Basic univariate and bivariate statistics for symbolic data: a critical review. Technical report
Klemelä J (2009) Smoothing of multivariate data: density estimation and visualization. Publisher: Wiley; 1 edition
Lauro CN, Gioia F (2006) Dependence and interdependence analysis for interval-valued variables. In: Bock H-H, Ferligoj A, Ziberna A (eds) Data Sci Classif, vol Batagelj. Springer-Verlag, Berlin, pp 171–183
Lauro CN, Palumbo F (2000) Principal component analysis of interval data: a symbolic analysis approach. Comput Stat 15(1):73–87
Lauro CN, Verde R, Irpino A (2008) Principal component analysis of symbolic data described by intervals, pp 279 – 311. In: Symbolic data analysis and the SODAS software edited by Edwin Diday. 2008
Lauro CN, Verde R (2000) Factorial data analysis on symbolic objects under cohesion constrains. In: Kiers HAL, Rasson JP, Groenen PJP, Schader M (eds) Data analysis classification and related methods. Springer-Verlag, Heidelberg
Le-Rademacher J, Billard L (2012) Symbolic-covariance principal component analysis and visualization for interval-valued data. J Comput Graph Stat 21(2):413–432
Lee JA, Verleysen M (2009) Quality assessment of dimensionality reduction: rank-based criteria. Neurocomputing 72:1431–1443
Li KC (1991) Sliced inverse regression for dimension reduction. J Am Stat Assoc 86:316–342
Li W, Guo J, Chen Y, Wang M (2016) A new representation of interval symbolic data and its application in dynamic clustering. J Classif 33(1):149–165
Liquet B, Saracco J (2012) A graphical tool for selecting the number of slices and the dimension of the model in SIR and SAVE approaches. Comput Stat 27:103–125
Lu HS, Wu HM (2010) Visualization, screening, and classification of cell cycle-regulated genes in yeast. Int J Syst Synth Biol 1(2):185–198
Maia ALS, de Carvalho FAT, Ludermir TB (2008) Forecasting models for interval-valued time series. Neurocomputing 71(16–18):3344–3352
Nueda MJ, Conesa A, Westerhuis JA, Hoefsloot HCJ, Smilde AK, Talon M, Ferrer A (2007) Discovering gene expression patterns in time course microarray experiments by ANOVA-SCA. Bioinformatics 23(14):1792–1800
Palumbo F, Lauro CN (2003) A PCA for interval valued data based on midpoints and radii. In: Yanai H, Okada A, Shigematu K, Kano Y, Meulman JJ (eds) New developments in psychometrics. Springer-Verlag, Japan, pp 641–648
Park J, Sriram TN, Yin X (2009) Central mean subspace in time series. J Comput Graph Stat 18:717–730
Park J, Sriram TN, Yin X (2010) Dimension reduction in time series. Stat Sinica 20:747–770
Raychaudhuri S, Stuart JM, Altman RB (2000) Principal components analysis to summarize microarray experiments: application to sporulation time series. Pacific symposium on biocomputing pp 455- 466
Sardá-Espinosa A (2019) Time-series clustering in r using the Dtwclust package. R J 11(1):22–43
Setodji CM, Cook RD (2004) K-means inverse regression. Technometrics 46(4):421–429
Spellman PT et al (1998) Comprehensive identification of cell cycle-regulated genes of the yeast saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell 9(12):3273–3297
Sun Y, Wong AKC, Kamel MS (2009) Classification of imbalanced data: a review. Int J Pattern Recogn Artif Intell 23(04):687–719
Teles P, Brito P (2015) Modeling interval time series with space-time. Commun Stat Theory Methods 44(17):3599–3627
Tsay RS (2010) Analysis of financial time series, 3rd edn. Wiley
Wall ME, Rechtsteiner A, Rocha LM (2003) Singular value decomposition and principal component analysis. In: A practical approach to microarray data analysis (D.P. Berrar, W. Dubitzky and M. Granzow, eds.) Kluwer: Norwell, MA, pp 91 – 109
Wang H, Guan R, Wu J (2012) CIPCA: complete-information-based principal component analysis for interval-valued data. Neurocomputing 86:158–169
Wei WWS (2019) Multivariate time series analysis with applications. Wiley
Wills G (2012) Visualizing time: designing graphical representations for statistical data (statistics and computing). Springer, Verlag New York Inc
Wu HM (2008) Kernel Sliced inverse regression with applications on classification. J Comput Graph Stat 17(3):590–610
Wu HM, Lu HHS (2004) Supervised motion segmentation by spatial-frequential analysis and dynamic sliced inverse regression. Stat Sinica 14:413–430
Wu HM, Kao CH, Chen CH (2020) Dimension reduction and visualization of symbolic interval-valued data using sliced inverse regression. In: Advances in data science: symbolic, complex, and network data (eds. Diday, E., Guan, R., Saporta, G., and Wang, H.). Wiley, pp 49 – 78
Yao WT, Wu HM (2013) Isometric sliced inverse regression or nonlinear manifolds learning. Stat Comput 23:563–576
Zhao J, Chevalier F, Pietriga E, Balakrishnan R (2011) Exploratory analysis of time-series with chronolenses. IEEE Transact Vis Comput Graph 17(12):2422–2431
Acknowledgements
We are grateful for the valuable comments provided by the Editor, Associate Editor, and anonymous referees. Their input has helped us improve the paper immensely.
Funding
This research was supported by grants from the Ministry of Science and Technology of Taiwan, R. O. C. under the grants MOST103-2118-M-032-006 and MOST111-2628-E-038-001-MY2.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Su, E.CY., Wu, HM. Dimension reduction and visualization of multiple time series data: a symbolic data analysis approach. Comput Stat 39, 1937–1969 (2024). https://doi.org/10.1007/s00180-023-01440-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00180-023-01440-7