Abstract
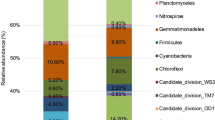
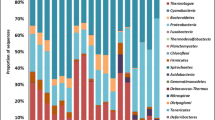
Microbial arsenic transformations play essential roles in controlling pollution and ameliorating risk. This study combined high-throughput sequencing and PCR-based approaches targeting both the 16 S rRNA and arsenic functional genes to investigate the temporal and spatial dynamics of the soil microbiomes impacted by high arsenic contamination (9.13 to 911.88 mg/kg) and to investigate the diversity and abundance of arsenic functional genes in soils influenced by an arsenic gradient. The results showed that the soil microbiomes were relatively consistent and mainly composed of Actinobacteria (uncultured Gaiellales and an unknown_67 − 14 bacterium), Proteobacteria, Firmicutes (particularly, Bacillus), Chloroflexi, and Acidobacteria (unknown_Subgroup_6). Although a range of arsenic functional genes (e.g., arsM, arsC, arrA, and aioA) were identified by shotgun metagenomics, only the arsM gene was detected by the PCR-based method. The relative abundance of the arsM gene accounted for 0.20%–1.57% of the total microbial abundance. Combining all analyses, arsenic methylation mediated by the arsM gene was proposed to be a key process involved in the arsenic biogeochemical cycle and mitigation of arsenic toxicity. This study advances our knowledge about arsenic mechanisms over the long-term in highly contaminated soils.


Similar content being viewed by others
References
Alotaibi BS, Khan M, Shamim S (2021) Unraveling the underlying heavy metal detoxification mechanisms of Bacillus species. Microorganisms 9:1628. https://doi.org/10.3390/microorganisms9081628
Andrews S (2010) FastQC: a quality control tool for high throughput sequence data. Available online http://www.bioinformatics.babraham.ac.uk/projects/fastqc
Bagade A, Nandre V, Paul D, Patil Y, Sharma N, Giri A, Kodam K (2020) Characterisation of hyper tolerant Bacillus firmus L-148 for arsenic oxidation. Environ Pollut 261:114124. https://doi.org/10.1016/j.envpol.2020.114124
Bose H, Sahu RP, Sar P (2022) Impact of arsenic on microbial community structure and their metabolic potential from rice soils of West Bengal, India. Sci Total Environ 841:156486. https://doi.org/10.1016/j.scitotenv.2022.156486
Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP (2016) DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. https://doi.org/10.1038/nmeth.3869
Chen S, Zhou Y, Chen Y, Gu J (2018) Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34:i884–i890. https://doi.org/10.1093/bioinformatics/bty560
Dong Y, Gao M, Liu X, Qiu W, Song Z (2020) The mechanism of polystyrene microplastics to affect arsenic volatilization in arsenic-contaminated paddy soils. J Hazard Mater 398:122896. https://doi.org/10.1016/j.jhazmat.2020.122896
Dunivin TK, Yeh SY, Shade A (2019) A global survey of arsenic-related genes in soil microbiomes. BMC Biol 17:1–17. https://doi.org/10.1186/s12915-019-0661-5
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. https://doi.org/10.1093/bioinformatics/btr381
Ewels P, Magnusson M, Lundin S, Käller M (2016) MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32:3047–3048. https://doi.org/10.1093/bioinformatics/btw354
Gong Y, Yang S, Chen S, Zhao S, Ai Y, Huang D, Yang K, Cheng H (2023) Soil microbial responses to simultaneous contamination of antimony and arsenic in the surrounding area of an abandoned antimony smelter in Southwest China. Environ Int 174:107897. https://doi.org/10.1016/j.envint.2023.107897
Huang JH (2014) Impact of microorganisms on arsenic biogeochemistry: a review. Water Air Soil Pollut 225:1848. https://doi.org/10.1007/s11270-013-1848-y
Jia Y, Huang H, Zhong M, Wang FH, Zhang LM, Zhu YG (2013) Microbial arsenic methylation in soil and rice rhizosphere. Environ Sci Technol 47:3141–3148. https://doi.org/10.1021/es303649v
Kandasamy S, Weerasuriya N, White JF, Patterson G, Lazarovits G (2021) Soil’s physical and nutritional balance is essential for establishing a healthy microbiome. In: Dubey SK, Sharma DK, Sharma JD (eds) Microbiome stimulants for crops. Woodhead Publishing, Cambridge, pp 381–404. https://doi.org/10.1016/B978-0-12-822122-8.00004-2
King KW, Williams MR, Macrae ML, Fausey NR, Frankenberger J, Smith DR, Kleinman PJA, Brown LC (2015) Phosphorus transport in agricultural subsurface drainage: a review. J Environ Qual 44:467–485. https://doi.org/10.2134/jeq2014.04.0163
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25:1754–1760. https://doi.org/10.1093/bioinformatics/btp324
Li Y, Zhang M, Xu R, Lin H, Sun X, Xu F, Gao P, Kong T, Xiao E, Yang N, Sun W (2021) Arsenic and antimony co-contamination influences on soil microbial community composition and functions: relevance to arsenic resistance and carbon, nitrogen, and sulfur cycling. Environ Int 153:106522. https://doi.org/10.1016/j.envint.2021.106522
Luo J, Bai Y, Liang J, Qu J (2014) Metagenomic approach reveals variation of microbes with arsenic and antimony metabolism genes from highly contaminated soil. PLOS ONE 9:e108185. https://doi.org/10.1371/journal.pone.0108185
Mirza BS, Sorensen DL, Dupont RR, McLean JE (2017) New arsenate reductase gene (arrA) PCR primers for diversity assessment and quantification in environmental samples. Appl Environ Microbiol 83:e02725–e02716. https://doi.org/10.1128/AEM.02725-16
Muller D, Lievremont D, Simeonova DD, Hubert JC, Lett MC (2003) Arsenite oxidase aox genes from a metal-resistant β-proteobacterium. J Bacteriol 185:135–141. https://doi.org/10.1128/JB.185.1.135-141.2003
Muyzer G, De Waal EC, Uitterlinden A (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695–700. https://doi.org/10.1128/aem.59.3.695-700.1993
Nurk S, Meleshko D, Korobeynikov A, Pevzner PA (2017) metaSPAdes: a new versatile metagenomic assembler. Genome Res 27:824–834. http://www.genome.org/cgi/doi/https://doi.org/10.1101/gr.213959.116
Qin J, Rosen BP, Zhang Y, Wang G, Franke S, Rensing C (2006) Arsenic detoxification and evolution of trimethylarsine gas by a microbial arsenite S-adenosylmethionine methyltransferase. Proc Natl Acad Sci 103:2075–2080. https://doi.org/10.1073/pnas.0506836103
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. https://doi.org/10.1093/nar/gks1219
Quéméneur M, Cébron A, Billard P, Battaglia-Brunet F, Garrido F, Leyval C, Joulian C (2010) Population structure and abundance of arsenite-oxidizing bacteria along an arsenic pollution gradient in waters of the Upper Isle River Basin, France. Appl Environ Microbiol 76:4566–4570. https://doi.org/10.1128/AEM.03104-09
Rahman Z, Thomas L, Singh VP (2020) Study of As-resistant bacteria from Nadia, India and a survey of two as resistance‐related proteins. J Basic Microbiol 60:47–57. https://doi.org/10.1002/jobm.201900373
Saltikov CW, Newman DK (2003) Genetic identification of a respiratory arsenate reductase. Proc Natl Acad Sci 100:10983–10988. https://doi.org/10.1073/pnas.1834303100
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. https://doi.org/10.1128/AEM.01541-09
Seemann T (2014) Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. https://doi.org/10.1093/bioinformatics/btu153
Sun Y, Polishchuk EA, Radoja U, Cullen WR (2004) Identification and quantification of arsC genes in environmental samples by using real-time PCR. J Microbiol Methods 58:335–349. https://doi.org/10.1016/j.mimet.2004.04.015
Tiankao W, Chotpantarat S (2018) Risk assessment of arsenic from contaminated soils to shallow groundwater in Ong Phra Sub-district, Suphan Buri Province, Thailand. J Hydrol-Reg Stud 19:80–96. https://doi.org/10.1016/j.ejrh.2018.08.001
Wright CH (1934) Soil analysis. A handbook of physical and chemical methods. Thomas Murby & Co., London
Xiao KQ, Li LG, Ma LP, Zhang SY, Bao P, Zhang T, Zhu YG (2016) Metagenomic analysis revealed highly diverse microbial arsenic metabolism genes in paddy soils with low-arsenic contents. Environ Pollut 211:1–8. https://doi.org/10.1016/j.envpol.2015.12.023
Yang X, Dai Z, Ge C, Yu H, Bolan N, Tsang DC, Song H, Hou D, Shaheen SM, Wang H, Rinklebe J (2023) Multiple-functionalized biochar affects rice yield and quality via regulating arsenic and lead redistribution and bacterial community structure in soils under different hydrological conditions. J Hazard Mater 443:130308. https://doi.org/10.1016/j.jhazmat.2022.130308
Yeager CM, Gallegos-Graves LV, Dunbar J, Hesse CN, Daligault H, Kuske CR (2017) Polysaccharide degradation capability of Actinomycetales soil isolates from a semiarid grassland of the Colorado Plateau. Appl Environ Microbiol 83:e03020–e03016. https://doi.org/10.1128/AEM.03020-16
Yi K, Li X, Chen D, Yang S, Liu Y, Tang X, Ling G, Zhao Z (2022) Shallower root spatial distribution induced by phosphorus deficiency contributes to topsoil foraging and low phosphorus adaption in sugarcane (Saccharum officinarum L). Front Plant Sci 12:3417. https://doi.org/10.3389/fpls.2021.797635
Zhai W, Wong MT, Luo F, Hashmi MZ, Liu X, Edwards EA, Tang X, Xu J (2017) Arsenic methylation and its relationship to abundance and diversity of arsM genes in composting manure. Sci Rep 7:42198. https://doi.org/10.1038/srep42198
Zhang SY, Zhao FJ, Sun GX, Su JQ, Yang XR, Li H, Zhu YG (2015) Diversity and abundance of arsenic biotransformation genes in paddy soils from southern China. Environ Sci Technol 49:4138–4146. https://doi.org/10.1021/acs.est.5b00028
Zhang SY, Xiao X, Chen SC, Zhu YG, Sun GX, Konstantinidis KT (2021) High arsenic levels increase activity rather than diversity or abundance of arsenic metabolism genes in paddy soils. Appl Environ Microbiol 87:e01383–e01321. https://doi.org/10.1128/AEM.01383-21
Zhao FJ, Harris E, Yan J, Ma J, Wu L, Liu W, McGrath SP, Zhou J, Zhu YG (2013) Arsenic methylation in soils and its relationship with microbial arsM abundance and diversity, and as speciation in rice. Environ Sci Technol 47:7147–7154. https://doi.org/10.1021/es304977m
Zhao C, Zhang Y, Chan Z, Chen S, Yang S (2015) Insights into arsenic multi-operons expression and resistance mechanisms in Rhodopseudomonas palustris CGA009. Front Microbiol 6:986. https://doi.org/10.3389/fmicb.2015.00986
Acknowledgements
This work (Grant No. RGNS 63–173) was supported by Office of the Permanent Secretary, Ministry of Higher Education, Science, Research and Innovation (OPS MHESI), Thailand Science Research and Innovation (TSRI) and Mahidol University. This research project was also supported by the Faculty of Science, Mahidol University. Philip D. Round provided helpful feedback and proofread the English of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sonthiphand, P., Rueangmongkolrat, N., Uthaipaisanwong, P. et al. Soil Microbiomes and their Arsenic Functional Genes in Chronically High-Arsenic Contaminated Soils. Bull Environ Contam Toxicol 112, 49 (2024). https://doi.org/10.1007/s00128-024-03866-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00128-024-03866-1