Abstract
Aims/hypothesis
Repeated exposures to insulin-induced hypoglycaemia in people with diabetes progressively impairs the counterregulatory response (CRR) that restores normoglycaemia. This defect is characterised by reduced secretion of glucagon and other counterregulatory hormones. Evidence indicates that glucose-responsive neurons located in the hypothalamus orchestrate the CRR. Here, we aimed to identify the changes in hypothalamic gene and protein expression that underlie impaired CRR in a mouse model of defective CRR.
Methods
High-fat-diet fed and low-dose streptozocin-treated C57BL/6N mice were exposed to one (acute hypoglycaemia [AH]) or multiple (recurrent hypoglycaemia [RH]) insulin-induced hypoglycaemic episodes and plasma glucagon levels were measured. Single-nuclei RNA-seq (snRNA-seq) data were obtained from the hypothalamus and cortex of mice exposed to AH and RH. Proteomic data were obtained from hypothalamic synaptosomal fractions.
Results
The final insulin injection resulted in similar plasma glucose levels in the RH group and AH groups, but glucagon secretion was significantly lower in the RH group (AH: 94.5±9.2 ng/l [n=33]; RH: 59.0±4.8 ng/l [n=37]; p<0.001). Analysis of snRNA-seq data revealed similar proportions of hypothalamic cell subpopulations in the AH- and RH-exposed mice. Changes in transcriptional profiles were found in all cell types analysed. In neurons from RH-exposed mice, we observed a significant decrease in expression of Avp, Pmch and Pcsk1n, and the most overexpressed gene was Kcnq1ot1, as compared with AH-exposed mice. Gene ontology analysis of differentially expressed genes (DEGs) indicated a coordinated decrease in many oxidative phosphorylation genes and reduced expression of vacuolar H+- and Na+/K+-ATPases; these observations were in large part confirmed in the proteomic analysis of synaptosomal fractions. Compared with AH-exposed mice, oligodendrocytes from RH-exposed mice had major changes in gene expression that suggested reduced myelin formation. In astrocytes from RH-exposed mice, DEGs indicated reduced capacity for neurotransmitters scavenging in tripartite synapses as compared with astrocytes from AH-exposed mice. In addition, in neurons and astrocytes, multiple changes in gene expression suggested increased amyloid beta (Aβ) production and stability. The snRNA-seq analysis of the cortex showed that the adaptation to RH involved different biological processes from those seen in the hypothalamus.
Conclusions/interpretation
The present study provides a model of defective counterregulation in a mouse model of type 2 diabetes. It shows that repeated hypoglycaemic episodes induce multiple defects affecting all hypothalamic cell types and their interactions, indicative of impaired neuronal network signalling and dysegulated hypoglycaemia sensing, and displaying features of neurodegenerative diseases. It also shows that repeated hypoglycaemia leads to specific molecular adaptation in the hypothalamus when compared with the cortex.
Data availability
The transcriptomic dataset is available via the GEO (http://www.ncbi.nlm.nih.gov/geo/), using the accession no. GSE226277. The proteomic dataset is available via the ProteomeXchange data repository (http://www.proteomexchange.org), using the accession no. PXD040183.
Graphical Abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.

Introduction
Repeated episodes of insulin-induced hypoglycaemia in individuals with type 1 or type 2 diabetes lead to defective secretion of glucagon and of the other counterregulatory hormones, adrenaline (epinephrine), noradrenaline (norepinephrine), cortisol and growth hormone [1]. Secretion of glucagon, which forms the first line of defence against hypoglycaemia, is controlled by multiple pancreatic-alpha-cell autonomous and non-autonomous mechanisms [2]. The role of central hypoglycaemia sensing in triggering glucagon secretion is well documented [3]. Neurons present in the brainstem, in particular in the dorsovagal complex, the parabrachial nucleus and the basolateral medulla, have been implicated in hypoglycaemia-induced glucagon secretion [4]. In the hypothalamus, glucose-sensing neurons activated by hypoglycaemia have been identified in the arcuate and in the paraventricular, the lateral and the dorsomedial nuclei. These neurons control glucagon secretion through modulation of the activity of the sympathetic or parasympathetic nerves and the sympatho-adrenal axis [3, 5]. In addition, magnocellular neurons of the paraventricular and supraoptic nuclei can be activated by hypoglycaemia, leading to secretion of arginine vasopressin (AVP) in the blood. AVP can then stimulate glucagon secretion by activating the AVP1b receptor in pancreatic islet alpha cells [6]. The mechanisms of hypoglycaemia sensing by neurons (referred to as glucose-inhibited [GI] neurons) are diverse but they share the ability to respond to a decrease in glucose metabolism and ATP production. This fall in intracellular energy level activates AMP-dependent protein kinase (AMPK), which controls membrane depolarisation by directly regulating the activity of ion channels and the production of reactive oxygen species [7, 8]. Alternatively, a reduction in the activity of the Na+/K+-ATPase induces membrane depolarisation and neuron firing [9, 10]. Thus, central hypoglycaemia sensing relies on a variety of signalling pathways, and there is little information about the dysegulations that impair the counterregulatory response (CRR) in people with diabetes treated with insulin.
Here, we sought to identify cellular and molecular pathways that are dysegulated in the hypothalamus of a mouse model of diabetes with defective hormonal counterregulation owing to exposure to single or repeated hypoglycaemic episodes.
Methods
Animals
Seven-week-old C57BL/6N male mice (Charles River Laboratories, Saint-Germain-Nuelles, France; http://www.criver.com/products-services/find-model/c57bl6-mouse?region=3616) were housed on a 12 h light/dark cycle. All procedures were approved by the Veterinary Office of Canton de Vaud (Switzerland; licence VD 3535) (see Electronic supplementary material [ESM] Methods). Blinding was not feasible during the generation of the model; however, results were analysed in a blinded fashion whenever possible.
High-fat diet and streptozocin treatment of mice and insulin-induced hypoglycaemia
Mice were fed a high-fat diet (HFD; 15.6% protein, 44.4% lipids, 40.3% carbohydrates; Diet 235HF, Safe, Paris, France) for 7 weeks. Subsequently, every 2 days, for approximately 10–14 days, after a 5 h fast, mice received i.p. injections of streptozocin (STZ; S0130, Sigma Aldrich, St Louis, MO, USA), twice at 35 mg/kg and once at 70 mg/kg (see ESM Methods, ‘High-fat diet and streptozocin treatment of mice’ section). Hypoglycaemia was induced by i.p. injection of insulin (Actrapid, Novo Nordisk, Bagsværd, Denmark). The recurrent hypoglycaemia (RH) group received three insulin injections per week for 4 weeks (12 injections in total). The acute hypoglycaemia (AH) group received three saline (154 mmol/l NaCl) injections per week for 3 weeks, followed by two further saline injections in week 4 plus a final injection of insulin. Plasma glucose was measured from tail-vein blood at −60 min, 60 min and 120 min after the final i.p. injection, using a glucometer (Contour XT, Bayer, Leverkusen, Germany). Plasma glucagon was measured from submandibular vein blood using an ELISA (catalogue no. 10-1271-01; Mercodia, Uppsala, Sweden) (see ESM Methods, ‘Insulin-induced hypoglycaemia and glucagon measurements’ for details).
Nuclei isolation, single-nuclei RNA-seq and data analysis
Brains were collected and snap frozen 120 min after the last insulin i.p. injection. Hypothalamus and cortex samples were dissected and nuclei were isolated (see ESM Methods, ‘Nuclei isolation’ section for details). For hypothalamus samples, single-nuclei libraries were prepared using the Chromium Next GEM Single Cell Multiome ATAC + Gene Expression assay (PN-1000283; 10x Genomics, Pleasanton, CA, USA), whilst, for cortex samples, we used the Chromium Single Cell 3′ RNA-seq assay (PN-1000075; 10x Genomics). All libraries were sequenced on the Illumina NovaSeq 6000 (Illumina, San Diego, CA, USA) (see ESM Methods, ‘Single-nuclei RNA sequencing’ section for details). The R package ‘Seurat’ (v4.1.1) [11] was used for data normalisation, quality control (QC) and clustering analysis (see ESM Methods, ‘Single-nuclei sequence analysis’ section). Gene ontology biological process (GO-BP) analysis and enrichment analysis of Kyoto Encyclopedia of Genes and Genomes (KEGG) terms were performed using a gene-set enrichment analysis (GSEA) approach using the R package ClusterProfiler (v4.4.4) [12] (see ESM Methods, ‘Gene-set enrichment analysis’ section).
Synaptosome proteomic analysis
Hypothalamic synaptosomal fractions were prepared following tissue homogenisation by differential centrifugation (see ESM Methods, ‘Synaptosome preparation’ section). Fractions were digested by trypsin and loaded on a TIMS-TOF Pro (Bruker, Bremen, Germany) mass spectrometer interfaced through a nanospray ion source to an Ultimate 3000 RSLCnano HPLC system (Dionex, Sunnyvale, CA, USA) (see ESM Methods, ‘Proteomics sample preparation’ and ‘LC-MS analyses’ sections). Data generation and analysis were performed as detailed in the ESM Methods, ‘Proteomics data analysis’ and ‘Bioinformatic analysis from proteomics data’ sections. All raw MS data, together with raw output tables, are available via the ProteomeXchange data repository (http://www.proteomexchange.org) with the accession PXD040183.
Statistical analysis
Unless stated otherwise, data are expressed as mean±SEM. Statistical analysis was performed using GraphPad Prism 8.4.0 (GraphPad Software, San Diego, CA, USA), using either a mixed-effects analysis followed by Sidak's post hoc test, a repeated-measures two-way ANOVA followed by Sidak's post hoc test, or an unpaired two-tailed Student's t test. Bonferroni and Benjamini–Hochberg corrections were applied for multiple comparisons. All p values <0.05 were considered to be significant.
Results
Repeated hypoglycaemia in mouse models of diabetes
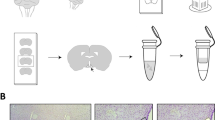
C57BL/6N mice were fed an HFD for 7 weeks and then received three STZ injections, as described in the methods (Fig. 1a), to create a mouse model of chemically induced type 2 diabetes. One week after the last STZ injection, the mice became hyperglycaemic and displayed lower body weight vs pre STZ treatment, although the latter change was non-significant (ESM Fig. 1a,b). Mice were divided into two groups of identical mean body weight and plasma glucose. One group received 11 injections of saline and a final injection of insulin over a period of 4 weeks (AH group) and the other group received 12 injections of insulin over the same period of time (RH group). The blood glucose levels before and after each injection are presented in ESM Fig. 1c. Plasma glucagon levels were measured 1 h after the last insulin injection. Insulin induced a deeper hypoglycaemia in the RH group (n=37) than in the AH group (n=33) (Fig. 1b; RH: 3.7±0.1 mmol/l; AH: 4.1±0.1 mmol/l; p=0.05) but a lower plasma glucagon response (Fig. 1c; RH: 59.0±4.8 ng/l; AH: 94.5±9.2 ng/l; ***p<0.001).
Recurrent exposure to hypoglycaemia reduces insulin-induced glucagon secretion. (a) Outline of the experiment to generate a model of type 2 diabetes and hypoglycaemia-associated autonomic failure (HAAF) in C57BL/6N male mice exposed to AH or RH. Created with BioRender.com. (b, c) Blood glucose (b) and plasma glucagon (c) in HFD-fed/STZ-treated mice exposed to AH or RH 1 h after a final i.p. injection of insulin (AH: n=33; RH: n=37). Data are means±SEM. ***p<0.001
Single-nuclei transcriptional profiling of the hypothalamus from AH- and RH-exposed diabetic mice
The hypothalami from three mice in the AH group and three mice in the RH group were collected 2 h after the last insulin injection and were pooled for each group for nuclei preparations and single-nuclei RNA-seq (snRNA-seq). The experiment was repeated once to obtain a biological replicate. We computed per-cell QC metrics to identify and remove low-quality nuclei, resulting in a dataset of 14,979 nuclei from the AH group and 14,934 nuclei from the RH group (ESM Fig. 2a–e). The major brain cell types could be identified based on the expression of their characteristic gene markers (Fig. 2a,b and ESM Fig. 3a). The cell type composition was very similar in the hypothalamus of AH and RH mice (Fig. 2c), with neurons being the most abundant, followed by oligodendrocytes and astrocytes. Microglia, endothelial cells and pericytes were much less represented and will not be discussed further.
Clustering and annotation of hypothalamic snRNA-seq in HFD-/STZ-treated mice exposed to RH and AH. (a) Two-dimensional Uniform Manifold Approximation and Projection (UMAP-1 and UMAP-2) representation of 14,979 and 14,934 nuclei isolated from the hypothalamus of AH- and RH-exposed mice, respectively. The cells are coloured based on the annotated cell type. (b) Violin plots of combined data from the RH and AH groups showing the expression of cellular-specific gene markers in each of the identified cell types. (c) Bar plot depicting the percentages of the different hypothalamic cell types per condition. Samples from n=3 mice were pooled for each group and the experiment was repeated once to obtain a biological replicate (n=2 integrated datasets per condition)
Differential hypothalamic gene expression analysis
Neurons
To delineate transcriptional changes occurring in the hypothalamus of mice in the RH vs AH group, we compared the transcriptional profiles for all major cell types. Analysis of neurons revealed that the majority of differentially expressed genes (DEGs) were downregulated after RH vs AH (Fig. 3a, Table 1 and ESM Table 1). Among the downregulated genes were Pcsk1n (an inhibitor of the proprotein convertase subtilisin/kexin type 1 [PCKS1]), Avp (encoding AVP, an inducer of glucagon secretion [6]) and Pmch (encoding pro-melanin concentrating hormone, an activator of the hypothalamic–pituitary–adrenal [HPA] axis) [13]. Other significantly downregulated genes were Resp18 [14], Rtn1 [15], Psap [16], Ubb [17], Aplp1 [18], Cst3 [19] and Itm2b [20], which are all associated with neurodegenerative diseases. Of note, Kcnq1ot1 [21], a long non-coding RNA (lncRNA) involved in diabetes susceptibility, was the only overexpressed gene.
Transcriptional analysis of neurons from hypothalami of HFD-/STZ-treated mice exposed to AH or RH. Samples from n=3 mice were pooled for each group and the experiment was repeated once to obtain a biological replicate (n=2 integrated datasets per condition). (a) Volcano plot depicting differential expression of 14,909 genes from hypothalamic neurons of mice subjected to RH as compared with AH. The red dot outlines the upregulated DEG and blue dots outline downregulated DEGs (fold change >1.2 or <−1.2 and Bonferroni adjusted p value [padj]<0.05). (b, c) Network visualisation of the top enriched GO-BP (b) or KEGG (c) terms in DEGs in the RH vs AH group. Node colour indicates the normalised enrichment score (NES); node size indicates the number of core-enriched genes; edges (grey lines) represent the pairwise similarity between terms
We next used GSEA to interrogate the GO-BP and KEGG databases to identify the biological processes regulated by the DEGs. Several GO-BP terms downregulated in the RH group were related to ‘oxidative phosphorylation’ (OXPHOS) and ‘ion homeostasis’ (Fig. 3b). Many OXPHOS genes were core-enriched in the ‘oxidative phosphorylation’ term, and genes encoding vacuolar and plasma membrane ATPases were present in the ‘ion homeostasis’ term (ESM Table 2). KEGG analysis revealed that the most downregulated terms were related to ‘neurodegenerative diseases’, which comprise OXPHOS genes belonging to Complexes I, III and IV and ATPsynthase, and ‘secretion activity’ comprising mostly the α and β subunits of the Na+/K+-ATPase, and several subunits of the vacuolar H+-ATPase (V-ATPase) (Fig. 3c and ESM Table 3).
GABAergic vs glutamatergic neurons
To gain further insight into neuron-specific transcriptional responses to RH, we analysed the two primary neuronal classes, GABAergic and glutamatergic neurons, identified by the expression of Gad1 and Gad2 genes, encoding γ-aminobutyric acid (GABA)-producing enzymes, or of the vesicular glutamate transporter-encoding gene Slc17a6, respectively (ESM Fig. 3b). Most of the DEGs (ESM Fig. 4a,b, Table 2 and ESM Table 1) were common to both neuronal populations. However, Avp and Pmch were significantly decreased in GABAergic neurons, as were two regulators of synaptic activity, Rabac1 [22] and Syt11 [23]. In glutamatergic neurons, there was decreased expression of Aldoa (a regulator of glycolysis [24]), Atp1a3 (encoding the α3 subunit of the Na+/K+-ATPase) and Ptprn (encoding protein tyrosine phosphatase N, which controls secretion of neurotransmitters [25]). There was also an increased expression of Dgkb (encoding diacylglycerol kinase, a regulator of dendritic outgrowth and spine maturation [26]). Thus, RH induced dysegulated expression of primarily the same genes in glutamatergic and GABAergic neurons, but with cell-specific changes in genes regulating synaptic vesicle secretion.
Oligodendrocytes
The volcano plot in Fig. 4a shows that there was reduced expression of Plp1, Mal, Scd2, Apoe, Ptgds, Cldn11 and Itm2b, which are all involved in various aspects of myelin formation [27], whilst the most overexpressed genes were Xylt1, Vcan and Zeb1 (ESM Table 1). GSEA analysis of DEGs revealed downregulation of GO-BP terms related to the ‘lipid biosynthesis’ and ‘myelination’ pathways (Fig. 4b, ESM Table 2). KEGG analysis showed significant increases in pathways related to various intracellular signalling pathways, including phosphoinositide 3 (PI3) kinases, protein kinase C and the cAMP pathway (Fig. 4c, ESM Table 3).
Transcriptional analysis of oligodendrocytes from hypothalami of HFD-/STZ-treated mice exposed to AH or RH. Samples from n=3 mice were pooled for each group and the experiment was repeated once to obtain a biological replicate (n=2 integrated datasets per condition). (a) Volcano plot depicting differential expression of 10,814 genes from hypothalamic oligodendrocytes of mice subjected to RH as compared with AH. Red dots outline upregulated DEGs and blue dots outline downregulated DEGs (fold change >1.2 or <−1.2 and Bonferroni adjusted p value [padj]<0.05); only genes with log2 fold change >0.5 or l<−0.5 have been labelled. (b, c) Network visualisation of the enriched GO-BP (b) or KEGG (c) terms in DEGs in RH- vs AH-exposed mice. Node colour indicates the normalised enrichment score (NES); node size indicates the number of core-enriched genes overlapping gene count; edges (grey lines) represent the pairwise similarity between terms. TRP, transient receptor potential
Astrocytes
GSEA analysis of DEGs (ESM Fig. 5a, Table 1 and ESM Table 1) showed repression of GO-BP terms related to ‘lipid biosynthesis’, ‘metabolism of amyloid beta protein’ and ‘ion transport’ (ESM Fig. 5b, ESM Table 2). Worth noting is the significant down-expression of Cst3, Apoe Itm2b, Clu and Atp6v0c (ESM Fig. 5a, Table 1), all involved in protecting against neurodegenerative diseases [19, 20, 28,29,30]. Similarly, a KEGG functional analysis showed decreased enrichment in pathways related to ‘neurodegenerative diseases’ and increased expression of ‘signalling pathways’ (ESM Fig. 5c, ESM Table 3).
Comparative analysis of transcriptomics and proteomic analysis
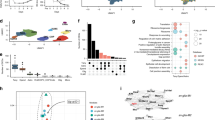
We performed proteomic analysis of synaptosomal fractions as our transcriptomic analysis indicated that several DEGs were related to synaptic structure and activity. Proteomic profiling of synaptosomal fractions from the hypothalamus of AH- and RH-exposed mice detected 7328 proteins in total, of which 6803 had their mRNAs detected in the snRNA-seq analysis (Fig. 5a). Comparative proteomics and transcriptomics GO-BP analysis identified several common terms related to ‘oxidative phosphorylation’ and ‘ion homeostasis’ (Fig. 5b, ESM Table 4). In these GO-BP terms, many proteins displayed changes in expression that correlated with changes in their mRNA expression (Fig. 5c). This indicates that transcript profiling is in large part predictive of changes in protein expression.
Comparison of neuronal transcriptomic and synaptosomal proteomic data from HFD-/STZ-treated mice exposed to AH or RH. Proteomics data originated from the analysis of the hypothalami of n=4 mice/group. (a) Venn diagram illustrating the number of features analysed in the proteomics analysis of synaptosomal fractions and in the transcriptomics analysis of neuronal nuclei from the hypothalamus of mice exposed to RH vs AH. (b) Hierarchical clustering of GO-BP terms enriched in both synaptosomal fractions and neuron transcriptomes of hypothalamus from RH- vs AH-exposed mice (|normalised enrichment score [NES]| >1.5 and Benjamini–Hochberg adjusted p value [padj]<0.05). Colour of cells indicates the NES. Corresponding padj are also indicated (*p<0.05, **p<0.01, ***p<0.001). (c) Heatmap depicting log2 fold change in expression and p values (*p<0.05, **p<0.01, ***p<0.001) of selected features linked to OXPHOS or ion homeostasis. (d) Volcano plot depicting differential expression of 7328 proteins detected in hypothalamic synaptosomes from mice subjected to RH as compared with AH. Red and blue dots indicate proteins that are significantly differentially expressed (fold change >2 or <−2 and p<0.05). ATP1B2, sodium/potassium-transporting ATPase subunit beta-2; ATP5A1/ATP5B/ATP5D/ATP5E/ATP5H/ATP5K/ATP5O, mitochondrial ATP synthase subunit alpha/beta/delta/epsilon/d/e/O; ATP5J, ATP synthase-coupling factor 6, mitochondrial; CORO1A, Coronin-1A; GRIN1, glutamate receptor; COX4I1/COX5B/COX7A2, mitochondrial cytochrome c oxidase subunit 4 isoform 1/subunit 5B/subunit 7A2; CYC1, cytochrome c1, heme protein, mitochondrial; NDUFA7/NDUFA8, NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8; NDUFB8, mitochondrial NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 8; NDUFB9, NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 9; NDUFC2, NADH dehydrogenase [ubiquinone] 1 subunit C2; NDUFS2/NDUFS7/NDUFS8, mitochondrial NADH dehydrogenase [ubiquinone] iron-sulfur protein 2/7/8; PRNP, major prion protein; SCO2, mitochondrial protein SCO2 homologue; SLC30A1, solute carrier family 30, member 1; SLC39A7, solute carrier family 39, member 7; SMDT1, single-pass membrane protein with aspartate rich tail 1; TMBIM6, Bax inhibitor motif containing 6; UQCC2, ubiquinol-cytochrome-c reductase complex assembly factor 2; UQCR10/UQCRB/QCRC1, cytochrome b-c1 complex subunit 9/7/1
Analysis of the synaptosomal proteins that showed highest differential expression between RH- and AH-exposed mice (Fig. 5d, Table 3), and for which their mRNA was not differentially expressed, revealed a relatively large group of proteins involved in synaptic remodelling [31, 32]. These included the downregulated proteins calcium/calmodulin dependent protein kinase II delta (CAMK2D), Rho guanine nucleotide exchange factor 7 (ARHGEF7), FYVE, also known as RhoGTP exchange factor and PH domain-containing protein 5 (FGD5), disintegrin and metalloproteinase domain-containing protein 22 (ADAM22) and Vang-like planar cell polarity protein 1 (VANGL1), and the upregulated proteins catenin delta-2 (CTNND2), Rho GTPase-activating protein 25 (ARHGAP25), USP6 N-terminal-like protein (USP6NL) and T cell lymphoma invasion and metastasis 2 (TIAM2) [31,32,33,34,35,36]. Other proteins found to be significantly differentially expressed are involved in mRNA splicing and export from the nucleus, including THO complex subunit 2 (THOC2), THO complex subunit 6 (THOC6) [37] and crooked neck pre-mRNA splicing factor 1 (CRNKL1) (all downregulated) and splicing factor, SR-related C-terminal domain associated factor 1 (SCAF1) (upregulated). Other upregulated proteins include the transcription factors double plant homeodomain fingers 2 (DPF2) and transducin-like enhancer family member 3, transcriptional corepressor (TLE3), and the signalling proteins segment polarity protein dishevelled segment polarity protein 2 (DVL2), protein phosphatase 1 regulatory inhibitor subunit 1B (PPP1R1B) and vasohibin 1 (VASH1). Since the mRNAs for these proteins were not found in the DEGs, this suggests that their expression is regulated at the post-translational level.
Single-nuclei transcriptional profiling of the cortex from AH and RH mice
To determine whether the impact of RH on the hypothalamus was specific for this brain structure, we repeated the snRNA-seq analysis in duplicate using nuclei from the cortex of RH- and AH-exposed mice. A total of 4650 and 9088 nuclei were obtained from the AH and RH groups, respectively (ESM Fig. 6a), and QC analysis was performed as for the hypothalamus (ESM Fig. 6b-e). Cluster annotation led to the identification of the same six cell types and similar relative distribution of these cells to that observed in the hypothalamus (ESM Fig. 7 and ESM Fig. 8a–c).
GO-BP analysis of DEGs in cortical neurons identified only a few enriched terms (ESM Fig. 9a,b, Table 4 and ESM Table 5). These were mostly related to ‘synapse organisation’ (ESM Fig. 9b, ESM Table 6) and only a few corresponded to those identified in the hypothalamus (Fig. 6a,b, and ESM Fig. 9) and often changed in the opposite direction in the two brain structures (Fig. 6b). Thus, RH induced different changes in gene expression in the hypothalamus and cortex, with more pronounced gene dysregulation in the hypothalamus.
Comparison of transcriptomic analysis from the hypothalamus and cortex of HFD-/STZ-treated mice exposed to AH or RH. For the hypothalamus, samples from n=3 mice were pooled for each group and the experiment was repeated once to obtain a biological replicate (n=2 integrated datasets per condition); a dataset of 14,979 nuclei from the AH group and 14,934 nuclei from the RH group was obtained. For the cortex, snRNA-seq analysis was conducted in duplicate using nuclei from the cortex of AH- and RH-exposed mice (samples from n=3 mice were pooled for each group) and a total of 4650 and 9088 nuclei were obtained, respectively. (a) Diagram representing the strategy followed. (b) Hierarchical clustering of GO-BP terms enriched in neuron transcriptomes (⎸normalised enrichment score [NES]⎹ >1.7) in the cortex and hypothalamus. Corresponding p values are also indicated (*p<0.05, **p<0.01, ***p<0.001)
Discussion
In this study, we investigated the modifications in hypothalamic gene and protein expression induced by repeated bouts of hypoglycaemia in a mouse model of diabetes with defective glucagon secretion. Using snRNA-seq analysis, we found that RH leads to important changes in gene expression in all the cell types discussed, with sufficient information obtained from neurons, oligodendrocytes and astrocytes for reliable differential gene expression analysis. Salient findings regarding the effect of RH include the reduced expression of Avp and Pmch, two neuropeptides involved in the CRR [6, 13]. In addition, there were changes suggestive of general defects in hypoglycaemia sensing, in synaptic activity and in neuron myelination; these changes are also associated with features of neurodegenerative diseases. Proteomic analysis of hypothalamic synaptosomal fractions further indicated that defects in synaptic activity were induced by RH. These results are summarised in Fig. 7. Finally, the hypothalamus appears much more sensitive to RH than the cortex.
Summary of the gene expression changes that may affect tripartite synapse function. In neurons, RH reduces the expression of OXPHOS genes (pink), of V- and Na+/K+-ATPase genes (brown), and of genes controlling neuropeptide expression, processing and synaptic vesicle exocytosis (violet). A decrease in OXPHOS-related gene expression reduces ATP production and, in combination with reduced expression of the Na+/K+-ATPase genes, overactivates GI neurons. In oligodendrocytes, many genes associated with myelination were downregulated, strongly suggesting a reduction in myelin formation capacity (yellow). In astrocytes, changes in gene expression suggest reduced neurotransmitter scavenging capacity (blue). In both neurons and astrocytes, several genes were downregulated that increased propensity for Aβ formation (green). NT, neurotransmitter. Created with BioRender.com
Neurons
Repeated hypoglycaemia mostly induced a reduction in gene expression. Among the most downregulated genes was Pcsk1n, an inhibitor of the protein convertase, PCSK1. This enzyme controls the functional maturation of several hormones and neuropeptides, including AVP, and the orexigenic and anorexigenic neuropeptides of the hypothalamic melanocortin pathway. By inactivating the proconvertase, the PCKS1 inhibitor (PCSK1N) regulates neuroendocrine secretory activity and has been associated with the development of diabetes [38]. PCSK1N is also considered as a potential target for the treatment of Alzheimer’s disease due to its anti-aggregant properties [39]. Thus, downregulation of Pcsk1n may have an impact on various aspects of the CRR through alterations of neuropeptide production and synaptic communication.
We also observed a strong decrease in the expression of Avp, which encodes the precursor of AVP, a hormone produced by magnocellular neurons of the paraventricular and supraoptic nuclei of the hypothalamus [6, 40]. It is secreted in response to hypoglycaemia and stimulates glucagon secretion. Reduced AVP secretion in individuals with type 1 diabetes explains part of their defective CRR [6]. Thus, AVP-producing neurons represent an important, yet vulnerable node in the integrated neuronal circuits that control the CRR.
In neurons, a single gene was upregulated by RH, Kcnq1ot1, an lncRNA, the overexpression of which inhibits the expression of various genes, and which has been linked to numerous complications of diabetes [21]. Whether Kcnq1ot1 overexpression is directly induced by hypoglycaemia and whether it contributes to the global changes in gene expression observed in our experiments will be interesting to further explore.
GO analysis of the differentially expressed neuronal genes revealed downregulation of the ‘oxidative phosphorylation’ pathway, with several genes associated with Complex I, Complex III, Complex IV and the ATP synthase being affected. These changes suggest a reduced capability to produce ATP following RH exposure. This will directly and negatively impact synaptic activity, a process that consumes a very large part of the ATP produced by mitochondria [41]. Defective mitochondrial function is also causally associated with dysregulated Ca2+ homeostasis and neurodegenerative diseases [42]. Several genes involved in amyloid production were also found to be dysregulated by RH, including Aplp [18], Rtn1 [15] and Itm2b [20], all of which have been associated with amyloid beta (Aβ) protein formation, and Resp18 [14], Psap [16], Ubb [17] and Cst3 [19], all of which are associated with neurodegenerative diseases.
In the ‘ion homeostasis’ GO terms, the most downregulated genes include Rnasek, Atp6v0b and Atp6v0c, which encode subunits of the V-ATPase, and Atp1b1, which encodes the β1 subunit of the Na+/K+-ATPase. The V-ATPase is required for synaptic vesicle acidification and trafficking and the Na+/K+-ATPase controls membrane potential [43]; both functions are essential for sustaining normal synaptic activity. Thus, repeated episodes of decreased energy availability caused by RH induce an energy-sparing state in neurons that is suggestive of reduced ATP production and decreased expression of ATPases. Collectively, these changes induce general impairment of synaptic activity.
Proteomic analysis of synaptosomal fractions
GO-BP analysis of proteomic data showed that the same ‘oxidative phosphorylation’ and ‘ion homeostasis’ terms that were identified in the transcriptomic analysis were downregulated by RH at the protein level. Many of the same genes and proteins associated with the OXPHOS Complex I, Complex III, Complex IV and the ATP synthase were downregulated by RH, as were those associated with the β subunit of the Na+/K+-ATPase. This indicates that a large number of the dysregulations observed at the mRNA level were reflected at the protein level. Functionally, this confirms that RH is likely to induce defective synaptic activity due to reduced OXPHOS activity and dysregulated control of membrane potential.
Among the proteins that were most differentially expressed in the synaptosomal fractions is a group of proteins involved in the control of synapse dynamics (see Fig. 5d). These included CAMK2D, a kinase activated by Ca2+ at excitatory synapses, which was downregulated by RH. CAMK2D activates ras-related C3 botulinum toxin substrate 1 (RAC1), a Rho-GTPase that triggers actin polymerisation, a critical process in the control of formation and function of spines and synapses [33]. This action of CAMK2D is mediated by phosphorylation of Rho-GTP exchange factors (GEFs) [34], which trigger the formation of active RAC1-GTP. Two of these Rho-GEFs, ARHGEF7 [44] and FGD5 [35], were downregulated in the RH mice and another, TIAM2, was upregulated. In contrast, among the proteins overexpressed after RH exposure are three Rho-GTPase-activating proteins (Rho-GAPs), CTNND2, ARHGAP25 and USP6NL, which increase GTP hydrolysis and RAC1 inactivation. These observations indicate important changes leading to reduced RAC1 activity. Interestingly, there was also decreased expression of ADAM22, a receptor for leucine-rich glioma-inactivated protein 1 beta (LGI1ß), which regulates synaptic maturation and activity [45]. Together, these observations indicate that RH leads to reduced formation of spines and synapses.
Oligodendrocytes
In oligodendrocytes, the major changes in gene expression induced by RH were related to ‘lipid biosynthesis’ and ‘myelination’, with decreased expression of the following: Plp1, the major constituent of myelin; Mal, which is associated with myelin formation; Scd2, which is the main stearoyl-CoA desaturase (SCD) isoform in the brain, the dysregulation of which is associated with impaired myelin formation and neurodegenerative diseases [46]; Apoe, implicated in brain lipid metabolism and neurodegeneration [28]; Ptgds which catalyses the formation of prostaglandin D2 (PGD2), a promoter of oligodendrocyte development and myelination [47]; and Cldn11, which is specifically required for the formation of myelin-associated tight junctions and the absence of which leads to major defects in neuron myelination [48]. Among the few upregulated mRNAs in oligodendrocytes from RH-exposed mice were Xylt1, required for heparan sulfate proteoglycan (HSPG) biosynthesis. HSPGs increase uptake of monomeric Tau protein, which induces an inflammatory response in neurons and astrocytes [49] and also blocks axon growth [50]. The other upregulated genes were Vcan, encoding a chondroitin sulfate proteoglycan that inhibits myelination of oligodendrocyte precursors [51], and Zeb1, a zinc finger transcription factor that induces neuroinflammation [52]. Together, these changes in oligodendrocyte gene expression indicate a generalised defect in myelin formation and the induction of local inflammation.
Astrocytes
In astrocytes, the most downregulated genes were Apoe, Itm2b, Itm2c, Cst3, Clu and Atp6v0c. Both Itm2b and Itm2c (a paralogue of Itm2b) are negative regulators of Aβ production, and Cst3 and Clu prevent aggregation of amyloid fibrils. Together, Itm2b, Itm2c, Cst3 and Clu potentially ameliorate Alzheimer’s disease [29, 53]. In addition, Atp6v0c encodes the pore-forming subunit of the V-ATPase, which is required to acidify lysosomes in which Aβ fibrils can be degraded [54].
The downregulated ‘ion transport’ GO-BP term is enriched in genes that encode voltage-gated K+ channels (Kcnj10, Kcnj2, Kcnh8), GABA receptors (Gabrg3, Gabbr1), the GABA transporter Slc6a1, and the glutamate transporter Slc1a1. These genes are required in astrocytes to clear neuronal synapses from excess neurotransmitters [55]. These changes in gene expression further suggest a generalised defect in tripartite synaptic activity and increased tendency to form Aβ peptides.
Cortex
Cortical neurons also displayed marked changes in gene expression in RH- vs AH-exposed mice. These were related mainly to presynapse organisation, with dysregulation in the synaptic regulators Nlgn1, Magi2, Il1rapl1, Nrxn1, Slitrk1, Lrfn5 and Mdga2 [56, 57]. However, the dysregulated genes observed in the cortex were very different than those observed in the hypothalamus of the same mice. Together, these findings indicate that the cortex is less and differently sensitive to RH than the hypothalamus.
Defective counterregulation
Defective counterregulation involves not only impaired glucagon secretion but also reduced secretion of adrenaline, noradrenaline, cortisol and growth hormones. These defects are thought to be caused by the progressive loss of central hypoglycaemia sensing and reduced activation of the autonomic nervous system, the HPA axis and AVP secretion. Mechanistically, activation of GI neurons by hypoglycaemia is triggered by a fall in intracellular ATP levels, which reduces the activity of the Na+/K+-ATPase, leading to membrane depolarisation and neuron firing [10]. For instance, a specific role of this signalling pathway in linking the activation of arcuate nucleus agouti-related peptide (AgRP) GI neurons to vagal nerve activity and glucagon secretion has been recently reported [58]. Thus, the observed decrease in neuronal expression of OXPHOS and Na+/K+-ATPase genes suggests over-activation of GI neurons in RH-exposed mice, which may also lead to their desensitisation to subsequent hypoglycaemic challenges. In addition, we observed decreased expression of Avp and of Pmch, which control glucagon [7] and corticosterone [13] secretion, respectively. Collectively, these changes in gene expression may not only explain basal hyperglucagonaemia in RH-exposed mice, but also the impaired response to subsequent hypoglycaemic episodes in animals with defective counterregulation.
Neurodegenerative diseases
Our data showed that several hypothalamic gene expression changes in RH- vs AH-exposed mice involved processes associated with neurodegenerative diseases, in particular Alzheimer’s disease, which is characterised by defects in energy homeostasis, synaptic and neuronal network dysfunction, and increased propensity for Aβ production (see Fig. 7) [59]. It is worth noting that snRNA-seq data showed that different modifications in gene expression was induced by RH in the cortex of the same mice. These were mostly related to synaptic activity and less so related to neurodegenerative pathways. These findings further indicate the specific, high sensitivity of the hypothalamus to the metabolic challenges imposed by RH.
Limitations of the study
The mouse model used has several limitations. First, our model of type 2 diabetes, with a degree of insulinopaenia, is more characteristic of long-term type 2 diabetes and does not fully represent the human condition. Second, our analysis did not identify whether the observed changes were specific to selected hypothalamic nuclei, or to GI neurons or neurons activated by high glucose (glucose-excited [GE] neurons). Also, our snRNA-seq analysis did not yield sufficient information on all hypothalamic cell types, therefore impairing the detection of potential relevant dysregulation in other cell types. Finally, whether the modifications in gene expression and glucagon secretion are reversible upon extended therapeutic restoration of normoglycaemia is not known.
In summary, we show that RH has an impact on all major hypothalamic cell types (Fig. 7). In neurons, it decreases the expression of OXPHOS genes and of plasma membrane and vacuolar ATPases that regulate synaptic activity, whilst in oligodendrocytes, RH downregulates the expression of genes controlling myelin formation and, in astrocytes, several genes controlling neurotransmitters scavenging at the synapse. In addition, many of the changes observed in neurons, oligodendrocytes and astrocytes suggest increased propensity for Aβ formation and accumulation. Importantly, defective CRR could be explained by the specific downregulation of Pcsk1, Avp and Pmch, and by the decreased expression of OXPHOS and Na+/K+-ATPase genes that may lead, not only to over-activation of hypoglycaemia-sensing neurons and increased basal hyperglucagonaemia, but also to impaired activation of these neurons during subsequent hypoglycaemic episodes. Together, our findings illustrate the very high sensitivity of the hypothalamus and its constituent cells to RH. They provide a framework to design novel experimental approaches to test the role of the identified pathways in defective counterregulation.
Change history
11 January 2024
A Correction to this paper has been published: https://doi.org/10.1007/s00125-023-06083-3
Abbreviations
- Aβ:
-
Amyloid beta
- ADAM22:
-
Disintegrin and metalloproteinase domain-containing protein 22
- AH:
-
Acute hypoglycaemia
- ARHGAP25:
-
Rho GTPase-activating protein 25
- ARHGEF7:
-
Rho guanine nucleotide exchange factor 7
- AVP:
-
Arginine vasopressin
- CAMK2D:
-
calcium/calmodulin dependent protein kinase II delta
- CRR:
-
Counterregulatory response
- CTNND2:
-
Catenin delta-2
- DEG:
-
Differentially expressed gene
- FGD5:
-
FYVE, RhoGEF and PH domain-containing protein 5
- GABA:
-
γ-Aminobutyric acid
- GEF:
-
GTP exchange factor
- GI:
-
Glucose-inhibited (neurons)
- GO:
-
Gene ontology
- GO-BP:
-
Gene ontology biological process
- GSEA:
-
Gene-set enrichment analysis
- HFD:
-
High-fat diet
- HPA:
-
Hypothalamic–pituitary–adrenal
- HSPG:
-
Heparan sulfate proteoglycan
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- lncRNA:
-
Long non-coding RNA
- OXPHOS:
-
Oxidative phosphorylation
- PCSK1:
-
Proprotein convertase subtilisin/kexin type 1
- PCKS1N:
-
Proprotein convertase subtilisin/kexin type 1 inhibitor
- QC:
-
Quality control
- RAC1:
-
Ras-related C3 botulinum toxin substrate 1
- RH:
-
Recurrent hypoglycaemia
- snRNA-seq:
-
Single-nuclei RNA-seq
- STZ:
-
Streptozocin
- TIAM2:
-
T cell lymphoma invasion and metastasis 2
- USP6NL:
-
USP6 N-terminal-like protein
- V-ATPase:
-
Vacuolar H+-ATPase
References
Cryer PE (2015) Hypoglycemia-associated autonomic failure in diabetes: maladaptive, adaptive, or both? Diabetes 64(7):2322–2323. https://doi.org/10.2337/db15-0331
MacDonald PE, Rorsman P (2023) Metabolic messengers: glucagon. Nat Metab 5(2):186–192. https://doi.org/10.1038/s42255-022-00725-3
Thorens B (2022) Neuronal regulation of glucagon secretion and gluconeogenesis. J Diabetes Investig 13(4):599–607. https://doi.org/10.1111/jdi.13745
Marty N, Dallaporta M, Thorens B (2007) Brain glucose sensing, counterregulation, and energy homeostasis. Physiology 22:241–251. https://doi.org/10.1152/physiol.00010.2007
Stanley S, Moheet A, Seaquist ER (2019) Central mechanisms of glucose sensing and counterregulation in defense of hypoglycemia. Endocr Rev 40(3):768–788. https://doi.org/10.1210/er.2018-00226
Kim A, Knudsen JG, Madara JC et al (2021) Arginine-vasopressin mediates counter-regulatory glucagon release and is diminished in type 1 diabetes. Elife 10:e72919. https://doi.org/10.7554/eLife.72919
Quenneville S, Labouèbe G, Basco D et al (2020) Hypoglycemia-sensing neurons of the ventromedial hypothalamus require AMPK-induced Txn2 expression but are dispensable for physiological counterregulation. Diabetes 69(11):2253–2266. https://doi.org/10.2337/db20-0577
Hirschberg PR, Sarkar P, Teegala SB, Routh VH (2020) Ventromedial hypothalamus glucose-inhibited neurones: a role in glucose and energy homeostasis? J Neuroendocrinol 32(1):e12773. https://doi.org/10.1111/jne.12773
Kurita H, Xu KY, Maejima Y et al (2015) Arcuate Na+, K+-ATPase senses systemic energy states and regulates feeding behavior through glucose-inhibited neurons. Am J Physiol Endocrinol Metab 309(4):E320–E333. https://doi.org/10.1152/ajpendo.00446.2014
Silver IA, Erecinska M (1998) Glucose-induced intracellular ion changes in sugar-sensitive hypothalamic neurons. J Neurophysiol 79(4):1733–1745. https://doi.org/10.1152/jn.1998.79.4.1733
Butler A, Hoffman P, Smibert P, Papalexi E, Satija R (2018) Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36(5):411–420. https://doi.org/10.1038/nbt.4096
Yu G, Wang LG, Han Y, He QY (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. Omics 16(5):284–287. https://doi.org/10.1089/omi.2011.0118
Kennedy AR, Todd JF, Dhillo WS et al (2003) Effect of direct injection of melanin-concentrating hormone into the paraventricular nucleus: further evidence for a stimulatory role in the adrenal axis via SLC-1. J Neuroendocrinol 15(3):268–272. https://doi.org/10.1046/j.1365-2826.2003.00997.x
Su J, Wang H, Yang Y et al (2018) RESP18 deficiency has protective effects in dopaminergic neurons in an MPTP mouse model of Parkinson’s disease. Neurochem Int 118:195–204. https://doi.org/10.1016/j.neuint.2018.06.010
Shi Q, Ge Y, He W, Hu X, Yan R (2017) RTN1 and RTN3 protein are differentially associated with senile plaques in Alzheimer’s brains. Sci Rep 7(1):6145. https://doi.org/10.1038/s41598-017-05504-9
Yoneshige A, Suzuki K, Suzuki K, Matsuda J (2010) A mutation in the saposin C domain of the sphingolipid activator protein (prosaposin) gene causes neurodegenerative disease in mice. J Neurosci Res 88(10):2118–2134. https://doi.org/10.1002/jnr.22371
Ryu KY, Garza JC, Lu XY, Barsh GS, Kopito RR (2008) Hypothalamic neurodegeneration and adult-onset obesity in mice lacking the Ubb polyubiquitin gene. Proc Natl Acad Sci U S A 105(10):4016–4021. https://doi.org/10.1073/pnas.0800096105
Guilarte TR (2010) APLP1, Alzheimer’s-like pathology and neurodegeneration in the frontal cortex of manganese-exposed non-human primates. Neurotoxicology 31(5):572–574. https://doi.org/10.1016/j.neuro.2010.02.004
Kaur G, Levy E (2012) Cystatin C in Alzheimer’s disease. Front Mol Neurosci 5:79. https://doi.org/10.3389/fnmol.2012.00079
Martins F, Santos I, da Cruz ESOAB, Tambaro S, Rebelo S (2021) The role of the integral type II transmembrane protein BRI2 in health and disease. Cell Mol Life Sci 78(21–22):6807–6822. https://doi.org/10.1007/s00018-021-03932-5
Xia F, Wang Y, Xue M et al (2022) LncRNA KCNQ1OT1: molecular mechanisms and pathogenic roles in human diseases. Genes Dis 9(6):1556–1565. https://doi.org/10.1016/j.gendis.2021.07.003
Fenster SD, Chung WJ, Zhai R et al (2000) Piccolo, a presynaptic zinc finger protein structurally related to bassoon. Neuron 25(1):203–214. https://doi.org/10.1016/S0896-6273(00)80883-1
Shimojo M, Madara J, Pankow S et al (2019) Synaptotagmin-11 mediates a vesicle trafficking pathway that is essential for development and synaptic plasticity. Genes Dev 33(5–6):365–376. https://doi.org/10.1101/gad.320077.118
Rose IA, O’Connell EL (1969) Studies on the interaction of aldolase with substrate analogues. J Biol Chem 244(1):126–134. https://doi.org/10.1016/S0021-9258(19)78201-5
Cai T, Fukushige T, Notkins AL, Krause M (2004) Insulinoma-associated protein IA-2, a vesicle transmembrane protein, genetically interacts with UNC-31/CAPS and affects neurosecretion in Caenorhabditis elegans. J Neurosci 24(12):3115–3124. https://doi.org/10.1523/JNEUROSCI.0101-04.2004
Hozumi Y, Watanabe M, Otani K, Goto K (2009) Diacylglycerol kinase beta promotes dendritic outgrowth and spine maturation in developing hippocampal neurons. BMC Neurosci 10:99. https://doi.org/10.1186/1471-2202-10-99
Depp C, Sun T, Sasmita AO et al (2023) Myelin dysfunction drives amyloid-β deposition in models of Alzheimer’s disease. Nature 618(7964):349–357. https://doi.org/10.1038/s41586-023-06120-6
Huang Y, Mahley RW (2014) Apolipoprotein E: structure and function in lipid metabolism, neurobiology, and Alzheimer’s diseases. Neurobiol Dis 72(Pt A):3–12. https://doi.org/10.1016/j.nbd.2014.08.025
Wojtas AM, Sens JP, Kang SS et al (2020) Astrocyte-derived clusterin suppresses amyloid formation in vivo. Mol Neurodegener 15(1):71. https://doi.org/10.1186/s13024-020-00416-1
Lo CH, Zeng J (2023) Defective lysosomal acidification: a new prognostic marker and therapeutic target for neurodegenerative diseases. Transl Neurodegener 12(1):29. https://doi.org/10.1186/s40035-023-00362-0
Saneyoshi T, Hayashi Y (2012) The Ca2+ and Rho GTPase signaling pathways underlying activity-dependent actin remodeling at dendritic spines. Cytoskeleton 69(8):545–554. https://doi.org/10.1002/cm.21037
Spence EF, Soderling SH (2015) Actin out: regulation of the synaptic cytoskeleton. J Biol Chem 290(48):28613–28622. https://doi.org/10.1074/jbc.R115.655118
Bai Y, Xiang X, Liang C, Shi L (2015) Regulating Rac in the nervous system: molecular function and disease implication of Rac GEFs and GAPs. Biomed Res Int 2015:632450. https://doi.org/10.1155/2015/632450
Saneyoshi T, Matsuno H, Suzuki A et al (2019) Reciprocal activation within a kinase-effector complex underlying persistence of structural LTP. Neuron 102(6):1199-1210 e1196. https://doi.org/10.1016/j.neuron.2019.04.012
Park S, Guo Y, Negre J et al (2021) Fgd5 is a Rac1-specific Rho GEF that is selectively inhibited by aurintricarboxylic acid. Small GTPases 12(2):147–160. https://doi.org/10.1080/21541248.2019.1674765
Fukata Y, Adesnik H, Iwanaga T, Bredt DS, Nicoll RA, Fukata M (2006) Epilepsy-related ligand/receptor complex LGI1 and ADAM22 regulate synaptic transmission. Science 313(5794):1792–1795. https://doi.org/10.1126/science.1129947
Maeder CI, Kim JI, Liang X et al (2018) The THO complex coordinates transcripts for synapse development and dopamine neuron survival. Cell 174(6):1436–1449. https://doi.org/10.1016/j.cell.2018.07.046
Ramos-Molina B, Martin MG, Lindberg I (2016) PCSK1 variants and human obesity. Prog Mol Biol Transl Sci 140:47–74. https://doi.org/10.1016/bs.pmbts.2015.12.001
Hoshino A, Helwig M, Rezaei S, Berridge C, Eriksen JL, Lindberg I (2014) A novel function for proSAAS as an amyloid anti-aggregant in Alzheimer’s disease. J Neurochem 128(3):419–430. https://doi.org/10.1111/jnc.12454
Gaspari S, Labouèbe G, Picard A, Berney X, Rodriguez Sanchez-Archidona A, Thorens B (2023) Tmem117 in AVP neurons regulates the counterregulatory response to hypoglycemia. EMBO Rep 24(8):e57344. https://doi.org/10.15252/embr.202357344
Harris JJ, Jolivet R, Attwell D (2012) Synaptic energy use and supply. Neuron 75(5):762–777. https://doi.org/10.1016/j.neuron.2012.08.019
Monzio Compagnoni G, Di Fonzo A, Corti S, Comi GP, Bresolin N, Masliah E (2020) The role of mitochondria in neurodegenerative diseases: the lesson from Alzheimer’s disease and Parkinson’s disease. Mol Neurobiol 57(7):2959–2980. https://doi.org/10.1007/s12035-020-01926-1
Pivovarov AS, Calahorro F, Walker RJ (2018) Na(+)/K(+)-pump and neurotransmitter membrane receptors. Invert Neurosci 19(1):1. https://doi.org/10.1007/s10158-018-0221-7
Manser E, Loo TH, Koh CG et al (1998) PAK kinases are directly coupled to the PIX family of nucleotide exchange factors. Mol Cell 1(2):183–192. https://doi.org/10.1016/s1097-2765(00)80019-2
Lovero KL, Fukata Y, Granger AJ, Fukata M, Nicoll RA (2015) The LGI1-ADAM22 protein complex directs synapse maturation through regulation of PSD-95 function. Proc Natl Acad Sci U S A 112(30):E4129-4137. https://doi.org/10.1073/pnas.1511910112
O’Neill LM, Guo CA, Ding F et al (2020) Stearoyl-CoA desaturase-2 in murine development, metabolism, and disease. Int J Mol Sci 21(22):8619. https://doi.org/10.3390/ijms21228619
Pan L, Trimarco A, Zhang AJ et al (2023) Oligodendrocyte-lineage cell exocytosis and L-type prostaglandin D synthase promote oligodendrocyte development and myelination. Elife 12:e77441. https://doi.org/10.7554/eLife.77441
Maheras KJ, Peppi M, Ghoddoussi F, Galloway MP, Perrine SA, Gow A (2018) Absence of claudin 11 in CNS myelin perturbs behavior and neurotransmitter levels in mice. Sci Rep 8(1):3798. https://doi.org/10.1038/s41598-018-22047-9
Song L, Oseid DE, Wells EA, Coaston T, Robinson AS (2022) Heparan sulfate proteoglycans (HSPGs) serve as the mediator between monomeric tau and its subsequent intracellular ERK1/2 pathway activation. J Mol Neurosci 72(4):772–791. https://doi.org/10.1007/s12031-021-01943-2
Hurtado A, Podinin H, Oudega M, Grimpe B (2008) Deoxyribozyme-mediated knockdown of xylosyltransferase-1 mRNA promotes axon growth in the adult rat spinal cord. Brain 131(Pt 10):2596–2605. https://doi.org/10.1093/brain/awn206
Ghorbani S, Jelinek E, Jain R et al (2022) Versican promotes T helper 17 cytotoxic inflammation and impedes oligodendrocyte precursor cell remyelination. Nat Commun 13(1):2445. https://doi.org/10.1038/s41467-022-30032-0
Poonaki E, Kahlert UD, Meuth SG, Gorji A (2022) The role of the ZEB1-neuroinflammation axis in CNS disorders. J Neuroinflammation 19(1):275. https://doi.org/10.1186/s12974-022-02636-2
Sastre M, Calero M, Pawlik M et al (2004) Binding of cystatin C to Alzheimer’s amyloid beta inhibits in vitro amyloid fibril formation. Neurobiol Aging 25(8):1033–1043. https://doi.org/10.1016/j.neurobiolaging.2003.11.006
Xiao Q, Yan P, Ma X et al (2014) Enhancing astrocytic lysosome biogenesis facilitates Abeta clearance and attenuates amyloid plaque pathogenesis. J Neurosci 34(29):9607–9620. https://doi.org/10.1523/JNEUROSCI.3788-13.2014
Sibille J, Pannasch U, Rouach N (2014) Astroglial potassium clearance contributes to short-term plasticity of synaptically evoked currents at the tripartite synapse. J Physiol 592(1):87–102. https://doi.org/10.1113/jphysiol.2013.261735
Sudhof TC (2017) Synaptic neurexin complexes: a molecular code for the logic of neural circuits. Cell 171(4):745–769. https://doi.org/10.1016/j.cell.2017.10.024
Goto-Ito S, Yamagata A, Sato Y et al (2018) Structural basis of trans-synaptic interactions between PTPdelta and SALMs for inducing synapse formation. Nat Commun 9(1):269. https://doi.org/10.1038/s41467-017-02417-z
Strembitska A, Labouebe G, Picard A et al (2022) Lipid biosynthesis enzyme Agpat5 in AgRP-neurons is required for insulin-induced hypoglycemia sensing and glucagon secretion. Nat Commun 13(1):5761. https://doi.org/10.1038/s41467-022-33484-6
Wilson DM III, Cookson MR, Van Den Bosch L, Zetterberg H, Holtzman DM, Dewachter I (2023) Hallmarks of neurodegenerative diseases. Cell 186(4):693–714. https://doi.org/10.1016/j.cell.2022.12.032
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Acknowledgements
The expert technical assistance of W. Dolci, X. P. Berney, S. Gaspari, A. Strembitska (University of Lausanne, Lausanne, Switzerland) and V. D. T. Tran (Swiss Insititute of Bioinformatics, Lausanne, Switzerland) is gratefully acknowledged. We thank the Genomic Technologies Facility of the University of Lausanne (Lausanne, Switzerland) for generating the snRNA-seq data and the Protein Analysis Facility from the University of Lausanne for the synaptosomal proteomic analysis. Some of the data were presented as an abstract at the 58th Annual Meeting of the EASD in 2022.
Data availability
All transcriptomic data are available via the GEO (http://www.ncbi.nlm.nih.gov/geo/), using the accession no. GSE226277. Proteomic data are available via the ProteomeXchange data repository (http://www.proteomexchange.org), using the accession no. PXD040183.
Funding
Open access funding provided by University of Lausanne This project has received funding from the Innovative Medicines Initiative 2 Joint Undertaking (JU), under grant agreement number 777460 (HypoRESOLVE). The JU receives support from the European Union’s Horizon 2020 research and innovation programme and the EFPIA and T1D Exchange, JDRF, International Diabetes Federation (IDF) and The Leona M and Harry B Helmsley Charitable Trust. The project was also supported by grants to BT from the Swiss National Science Foundation (310030–182496) from the European Research Council Advanced Grant (INTEGRATE, No. 694798).
Authors’ relationships and activities
JC-A was, and CF is employed by Novo Nordisk. The authors report no industry links or other personal connections. All other authors declare that there are no relationships or activities that might bias, or be perceived to bias, their work.
Contribution statement
JC-A participated in the study design, performed all the animal experimentations, prepared the nuclei for RNA-seq analysis and the synaptosomal fractions for proteomic analysis, analysed and interpreted the data, and wrote the paper. ARS-A participated in the study design, analysed the RNA-seq data and critically reviewed the paper. CF helped develop the mouse model, interpret the physiology data and corrected the manuscript. FM analysed the RNA-seq data, prepared the illustrations and wrote the paper. ME, AM and RJM contributed to the design of the experimental protocol and to the interpretation of the data, and critically reviewed the manuscript. MI contributed to the study design, data interpretation and writing the paper. BT conceived the study and supervised its execution, interpreted the data and wrote the paper. All authors commented on the paper and approved its final content. BT is the guarantor of this work.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised: The affiliations for Mark Evans, Alison McNeilly and Rory J. McCrimmon have been corrected.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Castillo-Armengol, J., Marzetta, F., Rodriguez Sanchez-Archidona, A. et al. Disrupted hypothalamic transcriptomics and proteomics in a mouse model of type 2 diabetes exposed to recurrent hypoglycaemia. Diabetologia 67, 371–391 (2024). https://doi.org/10.1007/s00125-023-06043-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-023-06043-x