Abstract
The type 2 diabetes epidemic and one of its predisposing factors, obesity, are major influences on global health and economic burden. It is accepted that genetics and the current environment contribute to this epidemic; however, in the last two decades, both human and animal studies have consolidated considerable evidence supporting the ‘developmental programming’ of these conditions, specifically by the intrauterine environment. Here, we review the various in utero exposures that are linked to offspring obesity and diabetes in later life, including epidemiological insights gained from natural historical events, such as the Dutch Hunger Winter, the Chinese famine and the more recent Quebec Ice Storm. We also describe the effects of gestational exposure to endocrine disruptors, maternal infection and smoking to the fetus in relation to metabolic programming. Causal evidence from animal studies, motivated by human observations, is also discussed, as well as some of the proposed underlying molecular mechanisms for developmental programming of obesity and type 2 diabetes, including epigenetics (e.g. DNA methylation and histone modifications) and microRNA interactions. Finally, we examine the effects of non-pharmacological interventions, such as improving maternal dietary habits and/or increasing physical activity, on the offspring epigenome and metabolic outcomes.
Similar content being viewed by others
Intrauterine exposures and programming of type 2 diabetes and obesity

Aside from the direct influences of genetics and the environment on an individual’s propensity to develop obesity and type 2 diabetes, the last three decades have seen strong evidence to support the notion that many adult-onset diseases are linked to in utero exposures. Hales and Barker proposed the ‘thrifty phenotype hypothesis’ to explain how poor availability of nutrients or a poor diet in utero results in poor fetal growth and programs structural and metabolic responses in the developing fetus [1]. These responses would be advantageous if the nutritional environment is reflected postnatally, but potentially deleterious in an energy-rich postnatal setting. This hypothesis has evolved to encompass the periconceptional period through to infancy and describes how adverse exposures that occur at critical points of development may affect function and/or structure of an organ system into adulthood; it is now referred to as the Developmental Origins of Health and Disease (DOHaD) hypothesis.
Fetal undernutrition and overnutrition
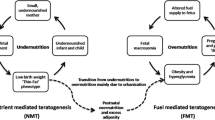
Fetal growth and birthweight are crude but commonly used measures of fetal wellbeing, shown to be regulated by maternal diet, lifestyle factors and the complex maternal–placental interplay [2]. Low birthweight is considered a marker for poor fetal nutritional status and has been associated with metabolic abnormalities, including type 2 diabetes and cardiovascular disease, in later life [3, 4]. Indeed, studies on prenatal famine during the Dutch Hunger Winter [5] showed that individuals exposed to famine while in utero exhibit decreased glucose tolerance some 50 years later compared with those born the year before the famine. Meanwhile, studies on adults born during the Chinese famine, between 1959 and 1961 [6], found that exposed individuals were more prone to be overweight and have type 2 diabetes, hyperglycaemia and the metabolic syndrome compared with those born after the famine. Additionally, interactions of the effects of the famine with an intergenerational risk of type 2 diabetes is cited as a major contributor to China’s current type 2 diabetes epidemic [7]. Twin studies support these findings: in monozygotic twin pairs discordant for type 2 diabetes, the twin with lower birthweight most often develops metabolic dysregulation [8]. Moreover, young adults with low birthweight display decreased muscle mass and height and increased fat mass compared with individuals of normal birthweight [4, 9]. Finally, more recent studies suggest that high birthweight is also associated with increased risk of obesity and type 2 diabetes [10]. This suggests that both fetal undernutrition and fetal overnutrition increase the risk of poor metabolic health later in life.
Fat and lean mass, both prenatally and in early postnatal life, also show relationships with in utero exposure, with a potential impact on future type 2 diabetes risk. For example, in a cohort of breastfeeding mother–infant dyads, in utero exposure to a higher maternal diet quality, based on the 2015 Healthy Eating Index (HEI-2015) [11], was inversely associated with infant body fat percentage [12]. Postnatally, breastfeeding or feeding a low-protein formula were associated with lower gain of fat mass (measured in children aged 5–8 years), whereas higher protein intake during the first 2 years postnatally resulted in higher BMI at 9 years of age and into adulthood [13].
Infections and inflammation
Metabolic and immune pathways are extensively integrated in health and disease. Specific metabolites in the cholesterol and tricarboxylic acid (TCA) cycle have an effect on inflammation [14, 15], and, conversely, infectious diseases in pregnancy may contribute to developmental origins of metabolic conditions [16]. Viral infections in pregnancy, specifically by enteroviruses [17], have been associated with type 1 diabetes in the offspring, though the mechanisms are complex and evidence circumstantial [18, 19]. Two recent systematic reviews and meta-analyses identified a potentially causative link between maternal viral infections in pregnancy and type 1 diabetes in the offspring [20, 21]. Interestingly, monocytes from mothers with gestational diabetes show a proinflammatory profile [22], which can also be induced in fetal monocytes of mothers infected with hepatitis B virus [22]. Together, these studies highlight a close relationship between hyperglycaemia and inflammatory memory [23]. Infections in pregnancy, such as premature births with chorioamnionitis, have been associated with histone modification changes in cord-blood monocytes [24], and inflammation memory in vitro is epigenetically modulated [25] and reversible [26]. These findings indicate that infection in utero can alter epigenetic patterns in offspring cells, supporting a causal link between infection and offspring obesity, mediated by metabolic and epigenetic reprogramming.
Environmental chemicals
Other prenatal exposures potentially linked to type 2 diabetes risk in later life include exposure to parental smoking [27, 28] and other environmental chemicals. For example, in utero exposure to dioxins, pesticides or bisphenol A in mice confers increased risk of developing type 2 diabetes [29]. In humans, exposure to organochlorines, as measured in second trimester maternal serum, was positively associated with BMI z scores and being overweight at 7 years of age [30]. Additionally, exposure to arsenic is linked to increased risk of gestational diabetes in the Maternal-Infant Research on Environmental Chemicals (MIREC) study [31] and in cohorts in France [32] and China [33], which poses an indirect threat to the affected offspring since gestational diabetes appears to be a programming factor for offspring metabolic dysfunction [34].
Maternal stress
Effects of prenatal maternal stress have been studied in natural disaster cohorts, such as Project Ice Storm, which included individuals who were exposed to the Quebec Ice Storm [35]. In the children of mothers who experienced hardship and stress during the ice storm, the severity of stress predicted the levels of insulin [36] and C-peptide [37] secretion. Similarly, a Danish longitudinal study found that children who were prenatally exposed to bereavement were more likely to have a type 2 diabetes diagnosis later in life [38]. Prenatal stress has also been shown to increase rat offspring susceptibility to diet-induced obesity [39]. Maternal sleep fragmentation-induced stress in mice has also been shown to result in offspring metabolic disorders, including increased body weight, visceral fat mass and HOMA-IR [40]. It is likely that future studies will continue to identify additional early-life exposures that impact the risk of later-life obesity and type 2 diabetes.
Gut microbiota
Disruption of the gut microbial community in newborns of obese mothers has also been shown to contribute to childhood inflammatory diseases, non-alcoholic fatty liver disease (NAFLD) and increased obesity risk [41]. This has been supported by studies showing that antibiotic use in the first year of life conferred an increased obesity risk [42], while synbiotics conferred protection against excessive fat accumulation under a high-fat diet (HFD) challenge [43]. In non-human primates, a maternal HFD was shown to reduce intestinal microbiota diversity in juvenile offspring at 1 year of age, even after switching to a healthy diet at the time of weaning [44].
Paternal factors
Until recently, programming research has focused mainly on maternal exposures to programming. Although limited, there is evidence in humans to support programming of type 2 diabetes and obesity via paternal exposures; a paradigm coined the Paternal Origins of Health and Disease (POHaD) [45]. For example, paternal smoking has been associated with increased body fat in male offspring [46], while paternal obesity is associated with type 1 diabetes in offspring [47]. Evidence from animal studies is much stronger and will be discussed later in this review.
Epigenetic mechanisms
Although the relationships between suboptimal in utero environments and increased risk of subsequent metabolic dysfunction are well established, underlying mechanisms have, until relatively recently, been poorly defined. In the last decade, numerous studies have implicated epigenetic mechanisms in the development of metabolic diseases through gene–environment interactions [48]. A range of exogenous exposures can influence epigenetic modifications, including the prenatal environment and adult lifestyle. Of particular note, compelling reproducible data have linked in utero exposure to smoking to defined changes in the offspring epigenome (see below).
Epigenetic mechanisms regulate gene activity in the absence of changes to the underlying DNA sequence, hence the name: ‘epi’, meaning ‘above’ in Greek, and ‘genetics’ [49]. Epigenetic mechanisms include DNA methylation, histone variants/modifications, chromatin-modifying proteins and non-coding RNAs. These processes regulate how densely specific regions of DNA are compacted, thus either inhibiting or enabling access of proteins, such as transcription factors, to DNA [50].
DNA methylation/demethylation
DNA methylation is the most studied epigenetic feature, primarily because its covalent chemical structure makes it highly stable and, therefore, quantifiable in a range of archived tissue and cells. DNA methylation is dispersed at varying densities across the genome, with specific variations of the methylation pattern being linked to cell identity and function [51]. In higher animal species, including humans, the main target is cytosines in CG dinucleotides, also referred to as CpG sites [52]. One feature of the vertebrate DNA methylation profile is the presence of CpG islands, regions of high-density CpG sites, located near or in gene-promoter regions. Around 29,000 CpG islands have been identified in the human genome [53]. DNA methylation in promoter regions may induce transcriptional inhibition or repression by affecting transcription-factor binding or recruiting proteins that specifically bind to methylated CpG sites [54].
DNA methyltransferases (DNMTs) transfer a methyl group to the 5′ position of cytosine. DNMT1, the maintenance methyltransferase, copies methylation status of hemimethylated sites after cell division [51]. In contrast DNMT3A and DNMT3B carry out de novo DNA methylation of unmethylated DNA, particularly in early embryonic development [55].
Demethylation can be a passive process, such as cell division without maintenance by DNMT1, or actively carried out by several enzymes, including the methylcytosine dioxygenases (ten-eleven translocation [TET]) enzymes that oxidise 5-methylcytosine (5meC) to 5-hydroxy methylcytosine (5hyroxy-meC) and other derivatives [56]. Each of these demethylation processes are important for appropriate gene expression and cell specification, particularly during early preimplantation development, as shown in Fig. 1 [57].
DNA methylation dynamics during human development. Male (blue line) and female (red line) embryos follow different DNA methylation patterns, from the birth of the parent through to zygote production (conception) and blastocyst implantation. Imprinted genes (dashed black line) do not undergo demethylation post-fertilisation and, hence, reflect parental-allele-specific methylation. PGC, primordial germ cells. Adapted from [125], with permission from Elsevier. This figure is available as part of a downloadable slideset
Although less dynamic, methylation also changes throughout postnatal life and adulthood. It is estimated that methylation of approximately 30% of all methylated sites in leucocytes or whole blood changes in an age-dependent manner [58, 59]. The methylation status of blood cells has also recently been shown to mirror age-related epigenetic signatures in adipose tissue [59]. Therefore, diet and other environmental factors throughout childhood and adulthood should also be considered when investigating epigenetic mechanisms in birth cohort studies of long-term health, since some epigenetic markers at specific loci appear to be much more flexible compared with those reported as stable markers over the long term [3].
The number of ‘epigenetic epidemiology’ papers and ‘epigenome-wide association studies’ (EWAS) published has increased sharply in the last 6 years and, coupled with locus- (gene-)specific epigenetic–environment studies, a plethora of data has emerged [60]. Despite good-practice approaches, such as publishing primer sequences and correctly referencing genome builds and CpG site locations, comparisons between datasets is not always straightforward, especially regarding the interpretation of what the ‘functional’ consequence of a change in DNA methylation means. For example, different quantification techniques have vastly different sensitivities and, therefore, some can detect small changes in DNA methylation (e.g. Infinium HumanMethylation arrays or targeted bisulphite sequencing), while others cannot (e.g. whole-genome bisulphite sequencing or bisulphite cloning and sequencing) [61]. Second, while genome-wide association studies (GWAS) studies can be carried out on any available cell type, DNA methylation varies between cell types and studies that use whole tissues or whole blood need to use algorithms to account for different cell types [62].
Histone modifications
Histone modifications occur in the N-terminal tail domains, in the core histone domains and in newly synthesised histones. Histone tails contain numerous sites that are amenable to acetylation and phosphorylation, which can alter the charge of the tails, thus affecting chromatin architecture through electrostatic mechanisms. These modifications act as ‘docking’ sites for chromatin ‘readers’ that recognise these modifications and recruit additional chromatin modifiers and remodelling enzymes [63]. It is now widely accepted that acetylation of histones inhibits the secondary and tertiary nucleosome structure, resulting in chromatin decondensation and increasing access to transcription factors and co-activators of transcription. In contrast, histone methylation has opposing effects, causing nucleosomal arrays to fold and condense, thus allowing active transcription [64].
microRNAs and long non-coding RNAs
Yet another regulatory mechanism contributing to phenotypic variation can occur at the post-transcriptional and transcriptional level; the emerging components of this type of regulation are microRNAs (miRNAs), which are small (21–24 nucleotide long) molecules that bind specifically to the 3′ untranslated regions of mRNA and interact with the Dicer complex. This binding sequesters the mRNA for degradation or prevents its translation by interfering with translation machinery. Additionally, long non-coding RNAs (lncRNAs) can bind mRNAs and act as molecular ‘sponges’ with opposing roles in transcript stabilisation/destabilisation. The roles of these two regulatory systems in type 2 diabetes pathogenesis has recently been reviewed by Saeedi et al [65].
Epigenetic variation in utero and metabolic programming
Maternal exposures
Several EWAS studies have found an association between maternal smoking and altered DNA methylation in cord blood [66], an effect that can persist postnatally [67] and into adulthood [68]. Maternal smoking has previously been linked to offspring obesity, with a linear dose-dependent effect, plateauing at 15 cigarettes or more per day [69]. Data from multiple studies and meta-analyses suggest a causative link between maternal smoking and increased risk of obese or overweight offspring [70]. Importantly DNA methylation at a specific gene, GFI1, was shown to mediate the effect of maternal smoking on offspring birthweight, explaining up to 19% of the difference in birthweight between offspring from mothers who smoked or did not smoke (control group) during pregnancy [71].
Maternal nutritional status and epigenetics
In the first study examining the effect of the Dutch Hunger Winter on epigenetic marks (i.e. chemical modifications on the DNA sequence), individuals who were 60 years old and prenatally exposed to this famine were found to have less DNA methylation at the imprinted IGF2 gene locus compared with their unexposed same-sex siblings [3]. More recently, genome-scale analysis in whole blood from this cohort identified that prenatal malnutrition-associated differentially methylated regions (P-DMRs) preferentially occurred at regulatory regions and were characterised by differential DNA methylation at regions associated with birthweight and serum LDL-cholesterol, i.e. INSR and CPT1A [72]. Hence, differential methylation of the P-DMRs extends along pathways related to growth and metabolism. Further exploratory analysis of six P-DMRs showed that they do not overlap with previously published adult tissue-specific differentially methylated regions (DMRs), highlighting that their establishment is dependent on specific exposure to famine during gestation.
Further evidence for a role of maternal nutrition in regulating the offspring epigenome comes from the Kiang West Longitudinal Population Study (KWLPS) [73], which included a cohort of 14,000 individuals from The Gambia that were subject to two distinct seasons, a hot dry ‘harvest’ season associated with high food abundance and a wet ‘hungry’ season associated with low energy intake [74]. Residents born in the hungry season were more likely to die prematurely (before the age of 25 years) [75] and to be small for gestational age [76]. Targeted epigenetic metastable epialleles, which are genomic regions that show significant inter-individual variation in DNA methylation in the absence of a genetic difference [77], were generally hypermethylated in individuals conceived during the hungry season, possibly as a result of increased one-carbon donor concentrations in the mother during this period [78]. Subsequent studies showed that multiple one-carbon donors, folate, riboflavin, betaine and choline all showed season-specific variation and their plasma concentrations predicted DNA methylation levels at metastable epialleles [79, 80]. The KWLPS cohort was used in conjunction with other datasets to identify a novel obesity-related (-predictive) metastable epiallele at the gene encoding pro-opiomelanocortin (POMC), which is similarly affected by maternal one-carbon donor concentration at conception [81].
Maternal overnutrition/obesity
The incidence of maternal obesity at conception and in pregnancy is increasing [82] and there is evidence that it contributes to increased infant birthweight (macrosomia and large for gestational age) and higher BMI in adolescent offspring [83, 84]. Excessive gestational weight gain (GWG) during pregnancy is also associated with increased offspring BMI and inflammatory markers (IL-6 and C-reactive protein), with early-gestation GWG having a stronger effect on offspring BMI at age 5 years and adiposity at age 9 years than mid–late-gestation GWG [85, 86]. Interestingly, while GWG in all trimesters affects birthweight, only first-trimester GWG affects child weight gain, suggesting that moderation during the first trimester may have the biggest impact on childhood weight [86].
Epigenetics are thought to mediate these effects, prompting several studies into the DNA methylation changes associated with maternal obesity [87,88,89]. Maternal diabetes correlates with obesity; in these studies, it was either removed as a covariate [88], was considered indistinguishable from obesity [87] or the cohort was structured to only contain pregnant women with obesity but not diabetes [88]. Two epigenome-wide studies analysed blood from the umbilical cord of offspring, and from 4–5-year-olds and 9–16-year-olds [87], who were exposed to maternal obesity (with or without gestational diabetes) and identified many differentially methylated sites in exposed offspring. Despite the relatively modest effect (generally <5% change), such data suggest that maternal obesity can lead to DNA methylation changes that are present at birth and remain postnatally.
Animal studies
Much of our understanding of programmed metabolic disease comes from animal models of under- and overnutrition. Studies in models of both ends of the nutritional spectrum have sought to understand potential programming mechanisms of type 2 diabetes and obesity risk by exploring epigenetic changes throughout the life course of exposed offspring. Importantly, unlike human studies, animal models allow the direct assessment of molecular and cellular defects.
Sperm and seminal fluid
Paternal low-protein diet (LPD) has been shown to enhance offspring fetal growth and predisposition to increased adiposity, glucose intolerance and cardiovascular dysfunction in the adult [90, 91], with both sperm and seminal fluid of LPD-fed fathers exerting programming effects. Similarly, diet-induced obesity in the father programs an impaired metabolic profile in his offspring, [92, 93]. In both fathers who were LPD and HFD fed [94], sperm cells displayed global DNA hypomethylation and altered miRNA expression. Aside from diet-induced programming, cold exposure has also been shown to induce differential methylation in sperm, which conferred enhanced brown adipose tissue (BAT) activity and protection from diet-induced obesity in male offspring [95]. In parallel, it was observed in humans that the presence of BAT and the season of conception were linked to offspring BMI.
Oocytes
Mitochondria are the most important organelle in the oocyte. While somatic cells maintain a healthy population of mitochondria by mitophagy, this pathway may not be active in oocytes. Thus, mitochondria damage in these germ cells may be transmitted to the developing blastocyst [96]. Such mitochondrial damage has been shown to occur in oocytes of obese dams, which have reduced mitochondrial DNA (mtDNA), accumulate the mitophagy marker phosphatase and tensin homologue-induced kinase 1 (PINK1) and demonstrate reduced developmental potential. The developing blastocysts show reduced levels of mtDNA and parallel mitochondrial loss in offspring that is caused by endoplasmic reticulum (ER) stress and which is reversible by ER stress inhibitors [97].
Pancreatic islets
Pancreatic failure and/or peripheral tissue insulin resistance are both programmed by adverse in utero exposures. Islet transcription factors are vulnerable to epigenetic changes as a response to suboptimal in utero environments leading to intrauterine growth restriction (IUGR). Uterine artery ligation in rats led to decreased histone H3 and H4 acetylation and loss-of-binding of the upstream stimulatory factor 1 (USF-1) transcription factor to the proximal promoter of Pdx1 in pancreatic islets, resulting in its markedly reduced transcription [98]. Maternal protein restriction in rats also led to reduced expression of Hnf4a in pancreatic islets of young male offspring in adulthood, which was associated with increased DNA methylation at the active Hnf4a promoter (P2) and increased repression through histone methylation at the enhancer region of this gene [99]. Consistently, the P2–enhancer interaction in islets of affected male offspring was significantly reduced, providing a mechanistic basis for reduced Hnf4a expression. Furthermore, the repressive histone mark, histone 3 lysine 27 trimethylation (H3K27me3), was found to accumulate with age in programmed offspring islets [99]. While insulin resistance was also observed in the female offspring in this model of IUGR, this was only evident in older mice [100]. Changes in DNA methylation have also been observed in pancreatic islets from a mouse model of maternal and fetal hyperglycaemia. Hypermethylation of the imprinted Igf2/H19 loci in pancreatic islets was observed and proposed to drive impaired islet structure and function [101] and, at the age studied, impaired glucose tolerance was more evident in male offspring than in females and accompanied by male-specific transmission to the next generation.
Adipose tissue
Adipose tissue has been shown to be an important target of developmental programming in animal models of both maternal undernutrition and overnutrition. In studies carried out only in male offspring and, specifically, in the epididymal white adipose tissue (eWAT), both maternal undernutrition [102] and maternal obesity [103] program an adipose tissue-insulin resistant phenotype accompanied by increased adiposity [102, 104, 105]. These programmed changes have both been attributed to epigenetic changes in adipose tissue. In addition, eWAT tissue hyperplasia due to maternal high-fat feeding during lactation was associated with increased expression and activity of stearoyl-CoA desaturase-1 (SCD1), a key enzyme in fatty acid metabolism. Changes in the expression of this enzyme were related to reduced DNA methylation of the Scd1 promoter [106].
Programmed changes in miRNAs have also been implicated in the programming of both adipose tissue expandability and insulin resistance. For example, using a rat model of maternal protein restriction, it has been shown that the imprinted miR-483 is programmed in eWAT of male offspring [107]. This was accompanied by a reduction in the expression of its direct target, Gdf3, and a reduction in the expandability of adipose tissue and, therefore, increased ectopic fat deposition, which is a major contributor to the development of insulin resistance. Importantly, an increase in adipose tissue miR-483 and parallel reduction in growth differentiation factor 3 (GDF-3) was also observed in adipose tissue from humans with low birthweight, showing conservation of this programmed mechanism. Programmed changes in miRNAs were also observed in a mouse model of maternal diet-induced obesity [103, 105]. Maternal feeding of a high-fat and high-simple-carbohydrate diet led to a programmed increase in miR-126, which led to a reduction in its direct target, insulin receptor substrate-1 (IRS-1), in eWAT of male offspring [103]. This programming effect was cell autonomous and was maintained in cultured pre-adipocytes differentiated in vitro, demonstrating that it was related to the programming of the adipocyte precursor stem cell pool.
Brain
The intrauterine environment also imposes important programming effects on the developing brain. Hypermethylation within a 500 bp region of the translation initiation start of the Pomc gene was observed in female offspring (Wistar outbred rats) exposed to maternal obesity in utero, corresponding with decreased Pomc transcription and increased body weight [108]. Diet-induced maternal obesity has also been shown to program feeding behaviour in the offspring by altering dopamine and opioid-related gene expression within the mesocorticolimbic reward pathways and hypothalamus [109]. This was linked to gene-specific promoter hypomethylation of the dopamine reuptake transporter, the μ-opioid receptor and proenkephalin, leading to an increased preference for sucrose and fat. The effects of overconsumption of these highly palatable and energy-dense foods are associated with obesity.
Conversely, in sheep, exposure to IUGR resulted in increased H3 lysine 9 acetylation (H3K9Ac) and decreased H3K27me3 modifications associated with the POMC promoter, and decreased methylation at a POMC proximal promoter region. However, these were not associated with either transcriptional or circulating POMC levels [110, 111]. In male C57BL/6J mice with IUGR followed by postnatal catch-up growth, differential expression and phosphorylation of components of the insulin signalling pathway in the arcuate nucleus of the hypothalamus effectively contributed to resistance to the anorectic effects of central insulin and impaired glucose homeostasis [112].
The importance of intervention studies
Lifestyle: diet and physical activity
The influence of dietary factors on both epigenetic patterns and phenotype provides a possible link between epigenetic marks and human metabolism.
Certain nutrients function as substrates for epigenetic modifications or co-factors for epigenetic enzymes and, therefore, diet can influence epigenetic patterns by varying epigenetic substrate availability or by altering the activity of enzymes that are involved in the addition or removal of epigenetic marks. A well-studied example is S-adenosyl methionine (SAM), a methyl donor substrate that provides methyl groups to both DNA and histone methyltransferases [113].
One of the strongest examples of an epigenetic alteration in adulthood, which is caused by environmental exposures during prenatal, childhood or adult life, is promoter DNA methylation of the key metabolic regulator, peroxisome proliferator-activated receptor, gamma, coactivator 1, alpha (PGC-1α) (encoded by PPARGC1A). PGC-1α plays a role in the regulation of genes required for energy metabolism, mitochondrial biogenesis and adaptive thermogenesis [114]. PPARGC1A expression is down-regulated in skeletal muscle from individuals that have impaired glucose tolerance or diabetes [115], while healthy men exposed to a high-fat overfeeding (HFO) diet for 5 days show increased DNA methylation at the PPARGC1A promoter in both adipose tissue and skeletal muscle [116, 117]. Feeding status has also been shown to affect methylation state; for example, 36 h of fasting affected DNA methylation of genes encoding leptin (LEP) and adiponectin (ADIPOQ) in adipose tissue [118].
Regular exercise has also been associated with wide-spread DNA methylation changes in a variety of tissues [119]. On the other hand, sedentary behaviour (9 days of bed rest) in healthy young men resulted in increased PPARGC1A DNA methylation and decreased PPARGC1A gene expression in skeletal muscle [120]. Recent data also shows that exercise may regulate histone deacetylases (HDAC) that further induce the expression of genes that play a role in metabolic pathways [121]. All in all, evidence suggests that a sedentary lifestyle can lead to genome-wide epigenetic changes and that physical exercise could be a possible mechanism to reverse these changes.
The benefits of increased physical activity have been interrogated in animal obesity or HFD-feeding models of programmed disease. In murine models of maternal obesity, daily treadmill running for 1 week prior to and throughout gestation led to improved insulin sensitivity in young adult offspring, which was associated with prevention of the programmed reduction in adipose tissue IRS-1 [122]. These exercise-driven improvements were analogous to those observed in 1-year-old offspring of mothers fed a HFD and housed with running wheels [123, 124]. The same research group showed similar benefits to 1-year-old offspring of HFD-fed fathers that had been exposed to voluntary exercise [92].
Conclusions and future perspectives
There is now compelling evidence for the transmission of poor metabolic health across generations. Mounting evidence shows that specific in utero environments (exposures) can have an impact on offspring epigenetic profile in a manner that is stable postnatally, into adulthood, in association with changed phenotype (Fig. 2). Despite these compelling data, only limited evidence exists for a causal role for epigenetic variation in mediating the effects of adverse in utero environment(s) on poor offspring metabolic health. Further additional longitudinal human studies are urgently needed to build this evidence base, supplemented with ongoing animal model studies that allow direct assessments of target tissues of relevance. Such a complementary approach should reveal the extent to which variation in epigenetic profile might act as a predictive early-life biomarker of increased metabolic risk, enabling targeting of novel interventions to those most likely to benefit. Further, the considerable interest in developing therapeutic epigenetic-modifying drugs and the increasing knowledge about the epigenetic-modifying properties of many dietary factors represent likely future approaches for modifying and reversing adverse metabolic health trajectories by (nutri) pharmacogenomic approaches.
A complex interplay between in utero environment, genetics, epigenetic marks and phenotype. (a) The fetus is exposed to various maternal exposures in utero, some of which are potentially detrimental, leading to activation of transcriptional regulators (via receptors) and of downstream genes. (b) Genetic differences also influence transcription factor binding and regulation of downstream gene expression (mRNA shown as ‘——AAA’). Transcription factor binding can lead to recruitment of epigenetic modifiers, reprogramming epigenetic marks at gene-regulatory elements. Variation in these epigenetic marks (e.g. DNA methylation, shown as grey shading of circles) often correlate with specific phenotypes at birth. (c) During postnatal life, individuals are continually exposed to environmental exposures that further modulate gene expression and phenotype. Differential epigenetic reprogramming in utero can affect the transcriptional response of cells to these new exposures, leading to different adult phenotypes. In the illustrated example, epigenetic remodelling of metabolism/nutrition genes during in utero development can lead to adult obesity. (d) A model showing how an initial exposure can alter the phenotype, leading to susceptibility to disease in response to a second environmental exposure. Solid lines indicate the order of events leading to phenotype B, while the dashed lines indicate the potential, but unproven, role for epigenetic variation in contributing to the phenotypes. The illustrations in (a–c) are based on decades of research showing that epigenetic marks can mediate the effect of exogenous signals on gene expression and phenotype. Nevertheless, showing causality is often difficult in humans and, in some cases, epigenetic differences may only correlate with exposure and outcome, but not necessarily contribute to phenotype. Longitudinal studies that make use of samples collected before disease onset are essential to tease out these causal/passenger questions. TF, transcription factor. This figure is available as part of a downloadable slideset
Abbreviations
- BAT:
-
Brown adipose tissue
- DNMT:
-
DNA methyltransferase
- ER:
-
Endoplasmic reticulum
- EWAS:
-
Epigenome-wide association studies
- eWAT:
-
Epididymal white adipose tissue
- GWG:
-
Gestational weight gain
- HFD:
-
High-fat diet
- H3K27me3:
-
Histone 3 lysine 27 trimethylation
- IUGR:
-
Intrauterine growth restriction
- KWLPS:
-
Kiang West Longitudinal Population Study
- LPD:
-
Low-protein diet
- miRNA:
-
MicroRNAs
- mtDNA:
-
Mitochondrial DNA
- P-DMR:
-
Prenatal malnutrition-associated differentially methylated region
- PGC-1α:
-
Peroxisome proliferator-activated receptor, gamma, coactivator 1, alpha
- POMC:
-
Pro-opiomelanocortin
References
Hales CN, Barker DJ (1992) Type 2 (non-insulin-dependent) diabetes mellitus: the thrifty phenotype hypothesis. Diabetologia 35(7):595–601. https://doi.org/10.1007/BF00400248
Fernandez-Twinn DS, Ozanne SE (2010) Early life nutrition and metabolic programming. Ann N Y Acad Sci 1212(1):78–96. https://doi.org/10.1111/j.1749-6632.2010.05798.x
Heijmans BT, Tobi EW, Stein AD et al (2008) Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci U S A 105(44):17046–17049. https://doi.org/10.1073/pnas.0806560105
Ravelli AC, van der Meulen JH, Michels RP et al (1998) Glucose tolerance in adults after prenatal exposure to famine. Lancet 351(9097):173–177. https://doi.org/10.1016/S0140-6736(97)07244-9
Lumey LH, Khalangot MD, Vaiserman AM (2015) Association between type 2 diabetes and prenatal exposure to the Ukraine famine of 1932-33: a retrospective cohort study. Lancet Diabetes Endocrinol 3(10):787–794. https://doi.org/10.1016/S2213-8587(15)00279-X
Li C, Lumey LH (2017) Exposure to the Chinese famine of 1959-61 in early life and long-term health conditions: a systematic review and meta-analysis. Int J Epidemiol 46(4):1157–1170. https://doi.org/10.1093/ije/dyx013
Zimmet P, Shi Z, El-Osta A, Ji L (2018) Epidemic T2DM, early development and epigenetics: implications of the Chinese Famine. Nat Rev Endocrinol 14(12):738–746. https://doi.org/10.1038/s41574-018-0106-1
Poulsen P, Vaag AA, Kyvik KO, Møller Jensen D, Beck-Nielsen H (1997) Low birth weight is associated with NIDDM in discordant monozygotic and dizygotic twin pairs. Diabetologia 40(4):439–446. https://doi.org/10.1007/s001250050698
Kensara OA, Wootton SA, Phillips DI et al (2005) Fetal programming of body composition: relation between birth weight and body composition measured with dual-energy X-ray absorptiometry and anthropometric methods in older Englishmen. Am J Clin Nutr 82(5):980–987. https://doi.org/10.1093/ajcn/82.5.980
Harder T, Rodekamp E, Schellong K, Dudenhausen JW, Plagemann A (2007) Birth weight and subsequent risk of type 2 diabetes: a meta-analysis. Am J Epidemiol 165(8):849–857. https://doi.org/10.1093/aje/kwk071
Krebs-Smith SM, Pannucci TE, Subar AF et al (2018) Update of the Healthy Eating Index: HEI-2015. J Acad Nutr Diet 118(9):1591–1602. https://doi.org/10.1016/j.jand.2018.05.021
Tahir MJ, Haapala JL, Foster LP et al (2019) Higher maternal diet quality during pregnancy and lactation is associated with lower infant weight-for-length, body fat percent, and fat mass in early postnatal life. Nutrients 11(3):632. https://doi.org/10.3390/nu11030632
Haschke F, Binder C, Huber-Dangl M, Haiden N (2019) Early-life nutrition, growth trajectories, and long-term outcome. Nestle Nutr Inst Workshop Ser 90:107–120. https://doi.org/10.1159/000490299
Arts RJ, Novakovic B, Ter Horst R et al (2016) Glutaminolysis and fumarate accumulation integrate immunometabolic and epigenetic programs in trained immunity. Cell Metab 24(6):807–819. https://doi.org/10.1016/j.cmet.2016.10.008
Bekkering S, Arts RJW, Novakovic B et al (2018) Metabolic induction of trained immunity through the mevalonate pathway. Cell 172(1–2):135–146. https://doi.org/10.1016/j.cell.2017.11.025
Bedu-Addo G, Alicke M, Boakye-Appiah JK et al (2017) In utero exposure to malaria is associated with metabolic traits in adolescence: The Agogo 2000 birth cohort study. J Inf Secur 75(5):455–463. https://doi.org/10.1016/j.jinf.2017.08.010
Dahlquist GG, Ivarsson S, Lindberg B, Forsgren M (1995) Maternal enteroviral infection during pregnancy as a risk factor for childhood IDDM. A population-based case-control study. Diabetes 44(4):408–413. https://doi.org/10.2337/diab.44.4.408
Filippi CM, von Herrath MG (2008) Viral trigger for type 1 diabetes: pros and cons. Diabetes 57(11):2863–2871. https://doi.org/10.2337/db07-1023
Coppieters KT, Boettler T, von Herrath M (2012) Virus infections in type 1 diabetes. Cold Spring Harb Perspect Med 2(1):a007682. https://doi.org/10.1101/cshperspect.a007682
Allen DW, Kim KW, Rawlinson WD, Craig ME (2018) Maternal virus infections in pregnancy and type 1 diabetes in their offspring: systematic review and meta-analysis of observational studies. Rev Med Virol 28(3):e1974. https://doi.org/10.1002/rmv.1974
Yue Y, Tang Y, Tang J et al (2018) Maternal infection during pregnancy and type 1 diabetes mellitus in offspring: a systematic review and meta-analysis. Epidemiol Infect 146(16):2131–2138. https://doi.org/10.1017/S0950268818002455
Yanai S, Tokuhara D, Tachibana D et al (2016) Diabetic pregnancy activates the innate immune response through TLR5 or TLR1/2 on neonatal monocyte. J Reprod Immunol 117:17–23. https://doi.org/10.1016/j.jri.2016.06.007
Netea MG, Joosten LA, Latz E et al (2016) Trained immunity: a program of innate immune memory in health and disease. Science 352(6284):aaf1098. https://doi.org/10.1126/science.aaf1098
Bermick J, Gallagher K, denDekker A, Kunkel S, Lukacs N, Schaller M (2019) Chorioamnionitis exposure remodels the unique histone modification landscape of neonatal monocytes and alters the expression of immune pathway genes. FEBS J 286(1):82–109. https://doi.org/10.1111/febs.14728
Foster SL, Hargreaves DC, Medzhitov R (2007) Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature 447(7147):972–978. https://doi.org/10.1038/nature05836
Novakovic B, Habibi E, Wang SY et al (2016) β-Glucan reverses the epigenetic state of LPS-induced immunological tolerance. Cell 167(5):1354–1368. https://doi.org/10.1016/j.cell.2016.09.034
Jaddoe VW, de Jonge LL, van Dam RM et al (2014) Fetal exposure to parental smoking and the risk of type 2 diabetes in adult women. Diabetes Care 37(11):2966–2973. https://doi.org/10.2337/dc13-1679
Montgomery SM, Ekbom A (2002) Smoking during pregnancy and diabetes mellitus in a British longitudinal birth cohort. BMJ 324(7328):26–27. https://doi.org/10.1136/bmj.324.7328.26
Alonso-Magdalena P, Quesada I, Nadal A (2011) Endocrine disruptors in the etiology of type 2 diabetes mellitus. Nat Rev Endocrinol 7(6):346–353. https://doi.org/10.1038/nrendo.2011.56
Agay-Shay K, Martinez D, Valvi D et al (2015) Exposure to endocrine-disrupting chemicals during pregnancy and weight at 7 years of age: a multi-pollutant approach. Environ Health Perspect 123(10):1030–1037. https://doi.org/10.1289/ehp.1409049
Ashley-Martin J, Dodds L, Arbuckle TE et al (2018) Association between maternal urinary speciated arsenic concentrations and gestational diabetes in a cohort of Canadian women. Environ Int 121(Pt 1):714–720. https://doi.org/10.1016/j.envint.2018.10.008
Marie C, Léger S, Guttmann A et al (2018) Exposure to arsenic in tap water and gestational diabetes: a French semi-ecological study. Environ Res 161:248–255. https://doi.org/10.1016/j.envres.2017.11.016
Xia X, Liang C, Sheng J et al (2018) Association between serum arsenic levels and gestational diabetes mellitus: a population-based birth cohort study. Environ Pollut 235:850–856. https://doi.org/10.1016/j.envpol.2018.01.016
Phelan S (2016) Windows of opportunity for lifestyle interventions to prevent gestational diabetes mellitus. Am J Perinatol 33(13):1291–1299. https://doi.org/10.1055/s-0036-1586504
King S, Laplante DP (2005) The effects of prenatal maternal stress on children’s cognitive development: Project Ice Storm. Stress 8(1):35–45. https://doi.org/10.1080/10253890500108391
Dancause KN, Veru F, Andersen RE, Laplante DP, King S (2013) Prenatal stress due to a natural disaster predicts insulin secretion in adolescence. Early Hum Dev 89(9):773–776. https://doi.org/10.1016/j.earlhumdev.2013.06.006
Cao-Lei L, Dancause KN, Elgbeili G, Laplante DP, Szyf M, King S (2018) DNA methylation mediates the effect of maternal cognitive appraisal of a disaster in pregnancy on the child’s C-peptide secretion in adolescence: Project Ice Storm. PLoS One 13(2):e0192199. https://doi.org/10.1371/journal.pone.0192199
Virk J, Li J, Vestergaard M, Obel C, Kristensen JK, Olsen J (2012) Prenatal exposure to bereavement and type-2 diabetes: a Danish longitudinal population based study. PLoS One 7(8):e43508. https://doi.org/10.1371/journal.pone.0043508
Tamashiro KL, Terrillion CE, Hyun J, Koenig JI, Moran TH (2009) Prenatal stress or high-fat diet increases susceptibility to diet-induced obesity in rat offspring. Diabetes 58(5):1116–1125. https://doi.org/10.2337/db08-1129
Trzepizur W, Khalyfa A, Qiao Z, Popko B, Gozal D (2017) Integrated stress response activation by sleep fragmentation during late gestation in mice leads to emergence of adverse metabolic phenotype in offspring. Metabolism 69:188–198. https://doi.org/10.1016/j.metabol.2017.01.026
Soderborg TK, Clark SE, Mulligan CE et al (2018) The gut microbiota in infants of obese mothers increases inflammation and susceptibility to NAFLD. Nat Commun 9(1):4462. https://doi.org/10.1038/s41467-018-06929-0
Azad MB, Bridgman SL, Becker AB, Kozyrskyj AL (2014) Infant antibiotic exposure and the development of childhood overweight and central adiposity. Int J Obes 38(10):1290–1298. https://doi.org/10.1038/ijo.2014.119
Mischke M, Arora T, Tims S et al (2018) Specific synbiotics in early life protect against diet-induced obesity in adult mice. Diabetes Obes Metab 20(6):1408–1418. https://doi.org/10.1111/dom.13240
Ma J, Prince AL, Bader D et al (2014) High-fat maternal diet during pregnancy persistently alters the offspring microbiome in a primate model. Nat Commun 5(1):3889. https://doi.org/10.1038/ncomms4889
Soubry A (2018) POHaD: why we should study future fathers. Environ Epigenet 4(2):dvy007. https://doi.org/10.1093/eep/dvy007
Northstone K, Golding J, Davey Smith G, Miller LL, Pembrey M (2014) Prepubertal start of father’s smoking and increased body fat in his sons: further characterisation of paternal transgenerational responses. Eur J Hum Genet 22(12):1382–1386. https://doi.org/10.1038/ejhg.2014.31
Magnus MC, Olsen SF, Granstrom C et al (2018) Paternal and maternal obesity but not gestational weight gain is associated with type 1 diabetes. Int J Epidemiol 47(2):417–426. https://doi.org/10.1093/ije/dyx266
Gluckman PD, Hanson MA, Buklijas T, Low FM, Beedle AS (2009) Epigenetic mechanisms that underpin metabolic and cardiovascular diseases. Nat Rev Endocrinol 5(7):401–408. https://doi.org/10.1038/nrendo.2009.102
Bird A (2007) Perceptions of epigenetics. Nature 447(7143):396–398. https://doi.org/10.1038/nature05913
Corpet A, Almouzni G (2009) Making copies of chromatin: the challenge of nucleosomal organization and epigenetic information. Trends Cell Biol 19(1):29–41. https://doi.org/10.1016/j.tcb.2008.10.002
Bird A (2002) DNA methylation patterns and epigenetic memory. Genes Dev 16(1):6–21. https://doi.org/10.1101/gad.947102
Ehrlich M, Gama-Sosa MA, Huang LH et al (1982) Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res 10(8):2709–2721. https://doi.org/10.1093/nar/10.8.2709
Lander ES, Linton LM, Birren B et al (2001) Initial sequencing and analysis of the human genome. Nature 409(6822):860–921. https://doi.org/10.1038/35057062
Ling C, Groop L (2009) Epigenetics: a molecular link between environmental factors and type 2 diabetes. Diabetes 58(12):2718–2725. https://doi.org/10.2337/db09-1003
Okano M, Bell DW, Haber DA, Li E (1999) DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99(3):247–257. https://doi.org/10.1016/S0092-8674(00)81656-6
Rasmussen KD, Helin K (2016) Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev 30(7):733–750. https://doi.org/10.1101/gad.276568.115
Guo F, Li X, Liang D et al (2014) Active and passive demethylation of male and female pronuclear DNA in the mammalian zygote. Cell Stem Cell 15(4):447–459. https://doi.org/10.1016/j.stem.2014.08.003
Hannum G, Guinney J, Zhao L et al (2013) Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell 49(2):359–367. https://doi.org/10.1016/j.molcel.2012.10.016
Rönn T, Volkov P, Gillberg L et al (2015) Impact of age, BMI and HbA1c levels on the genome-wide DNA methylation and mRNA expression patterns in human adipose tissue and identification of epigenetic biomarkers in blood. Hum Mol Genet 24(13):3792–3813. https://doi.org/10.1093/hmg/ddv124
Foley DL, Craig JM, Morley R et al (2009) Prospects for epigenetic epidemiology. Am J Epidemiol 169(4):389–400. https://doi.org/10.1093/aje/kwn380
Bock C, Halbritter F, Carmona FJ et al (2016) Quantitative comparison of DNA methylation assays for biomarker development and clinical applications. Nat Biotechnol 34(7):726–737. https://doi.org/10.1038/nbt.3605
Houseman EA, Accomando WP, Koestler DC et al (2012) DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 13(1):86. https://doi.org/10.1186/1471-2105-13-86
Gillette TG, Hill JA (2015) Readers, writers, and erasers: chromatin as the whiteboard of heart disease. Circ Res 116(7):1245–1253. https://doi.org/10.1161/CIRCRESAHA.116.303630
Tessarz P, Kouzarides T (2014) Histone core modifications regulating nucleosome structure and dynamics. Nat Rev Mol Cell Biol 15(11):703–708. https://doi.org/10.1038/nrm3890
Saeedi Borujeni MJ, Esfandiary E, Baradaran A et al (2019) Molecular aspects of pancreatic β-cell dysfunction: oxidative stress, microRNA, and long noncoding RNA. J Cell Physiol 234(6):8411–8425. https://doi.org/10.1002/jcp.27755
Joubert BR, Håberg SE, Nilsen RM et al (2012) 450K epigenome-wide scan identifies differential DNA methylation in newborns related to maternal smoking during pregnancy. Environ Health Perspect 120(10):1425–1431. https://doi.org/10.1289/ehp.1205412
Novakovic B, Ryan J, Pereira N, Boughton B, Craig JM, Saffery R (2014) Postnatal stability, tissue, and time specific effects of AHRR methylation change in response to maternal smoking in pregnancy. Epigenetics 9(3):377–386. https://doi.org/10.4161/epi.27248
Richmond RC, Simpkin AJ, Woodward G et al (2015) Prenatal exposure to maternal smoking and offspring DNA methylation across the lifecourse: findings from the Avon Longitudinal Study of Parents and Children (ALSPAC). Hum Mol Genet 24(8):2201–2217. https://doi.org/10.1093/hmg/ddu739
Albers L, Sobotzki C, Kuß O et al (2018) Maternal smoking during pregnancy and offspring overweight: is there a dose-response relationship? An individual patient data meta-analysis. Int J Obes 42(7):1249–1264. https://doi.org/10.1038/s41366-018-0050-0
Behl M, Rao D, Aagaard K et al (2013) Evaluation of the association between maternal smoking, childhood obesity, and metabolic disorders: a national toxicology program workshop review. Environ Health Perspect 121(2):170–180. https://doi.org/10.1289/ehp.1205404
Küpers LK, Xu X, Jankipersadsing SA et al (2015) DNA methylation mediates the effect of maternal smoking during pregnancy on birthweight of the offspring. Int J Epidemiol 44(4):1224–1237. https://doi.org/10.1093/ije/dyv048
Tobi EW, Goeman JJ, Monajemi R et al (2014) DNA methylation signatures link prenatal famine exposure to growth and metabolism. Nat Commun 5(1):5592. https://doi.org/10.1038/ncomms6592
Hennig BJ, Unger SA, Dondeh BL et al (2017) Cohort profile: the Kiang West Longitudinal Population Study (KWLPS)—a platform for integrated research and health care provision in rural Gambia. Int J Epidemiol 46(2):e13. https://doi.org/10.1093/ije/dyv206
Moore SE (2017) Early-life nutritional programming of health and disease in The Gambia. Ann Nutr Metab 70(3):179–183. https://doi.org/10.1159/000456555
Moore SE, Cole TJ, Poskitt EM et al (1997) Season of birth predicts mortality in rural Gambia. Nature 388(6641):434. https://doi.org/10.1038/41245
Rayco-Solon P, Fulford AJ, Prentice AM (2005) Differential effects of seasonality on preterm birth and intrauterine growth restriction in rural Africans. Am J Clin Nutr 81(1):134–139. https://doi.org/10.1093/ajcn/81.1.134
Rakyan VK, Blewitt ME, Druker R, Preis JI, Whitelaw E (2002) Metastable epialleles in mammals. Trends Genet 18(7):348–351. https://doi.org/10.1016/S0168-9525(02)02709-9
Waterland RA, Kellermayer R, Laritsky E et al (2010) Season of conception in rural gambia affects DNA methylation at putative human metastable epialleles. PLoS Genet 6(12):e1001252. https://doi.org/10.1371/journal.pgen.1001252
Dominguez-Salas P, Moore SE, Cole D et al (2013) DNA methylation potential: dietary intake and blood concentrations of one-carbon metabolites and cofactors in rural African women. Am J Clin Nutr 97(6):1217–1227. https://doi.org/10.3945/ajcn.112.048462
Dominguez-Salas P, Moore SE, Baker MS et al (2014) Maternal nutrition at conception modulates DNA methylation of human metastable epialleles. Nat Commun 5(1):3746. https://doi.org/10.1038/ncomms4746
Kühnen P, Handke D, Waterland RA et al (2016) Interindividual variation in DNA methylation at a putative POMC metastable epiallele is associated with obesity. Cell Metab 24(3):502–509. https://doi.org/10.1016/j.cmet.2016.08.001
Poston L, Caleyachetty R, Cnattingius S et al (2016) Preconceptional and maternal obesity: epidemiology and health consequences. Lancet Diabetes Endocrinol 4(12):1025–1036. https://doi.org/10.1016/S2213-8587(16)30217-0
Gaillard R, Welten M, Oddy WH et al (2016) Associations of maternal prepregnancy body mass index and gestational weight gain with cardio-metabolic risk factors in adolescent offspring: a prospective cohort study. BJOG 123(2):207–216. https://doi.org/10.1111/1471-0528.13700
Yu Z, Han S, Zhu J, Sun X, Ji C, Guo X (2013) Pre-pregnancy body mass index in relation to infant birth weight and offspring overweight/obesity: a systematic review and meta-analysis. PLoS One 8(4):e61627. https://doi.org/10.1371/journal.pone.0061627
Fraser A, Tilling K, Macdonald-Wallis C et al (2010) Association of maternal weight gain in pregnancy with offspring obesity and metabolic and vascular traits in childhood. Circulation 121(23):2557–2564. https://doi.org/10.1161/CIRCULATIONAHA.109.906081
Margerison-Zilko CE, Shrimali BP, Eskenazi B, Lahiff M, Lindquist AR, Abrams BF (2012) Trimester of maternal gestational weight gain and offspring body weight at birth and age five. Matern Child Health J 16(6):1215–1223. https://doi.org/10.1007/s10995-011-0846-1
Hjort L, Martino D, Grunnet LG et al (2018) Gestational diabetes and maternal obesity are associated with epigenome-wide methylation changes in children. JCI Insight 3(17):e122572. https://doi.org/10.1172/jci.insight.122572
Martin CL, Jima D, Sharp GC et al (2019) Maternal pre-pregnancy obesity, offspring cord blood DNA methylation, and offspring cardiometabolic health in early childhood: an epigenome-wide association study. Epigenetics 14(4):325–340. https://doi.org/10.1080/15592294.2019.1581594
Nogues P, Dos Santos E, Jammes H et al (2019) Maternal obesity influences expression and DNA methylation of the adiponectin and leptin systems in human third-trimester placenta. Clin Epigenetics 11(1):20. https://doi.org/10.1186/s13148-019-0612-6
Watkins AJ, Sinclair KD (2014) Paternal low protein diet affects adult offspring cardiovascular and metabolic function in mice. Am J Physiol Heart Circ Physiol 306(10):H1444–H1452. https://doi.org/10.1152/ajpheart.00981.2013
Watkins AJ, Dias I, Tsuro H et al (2018) Paternal diet programs offspring health through sperm- and seminal plasma-specific pathways in mice. Proc Natl Acad Sci U S A 115(40):10064–10069. https://doi.org/10.1073/pnas.1806333115
Stanford KI, Rasmussen M, Baer LA et al (2018) Paternal exercise improves glucose metabolism in adult offspring. Diabetes 67(12):2530–2540. https://doi.org/10.2337/db18-0667
Huypens P, Sass S, Wu M et al (2016) Epigenetic germline inheritance of diet-induced obesity and insulin resistance. Nat Genet 48(5):497–499. https://doi.org/10.1038/ng.3527
Fullston T, Ohlsson Teague EM, Palmer NO et al (2013) Paternal obesity initiates metabolic disturbances in two generations of mice with incomplete penetrance to the F2 generation and alters the transcriptional profile of testis and sperm microRNA content. FASEB J 27(10):4226–4243. https://doi.org/10.1096/fj.12-224048
Sun W, Dong H, Becker AS et al (2018) Cold-induced epigenetic programming of the sperm enhances brown adipose tissue activity in the offspring. Nat Med 24(9):1372–1383. https://doi.org/10.1038/s41591-018-0102-y
Boudoures AL, Saben J, Drury A et al (2017) Obesity-exposed oocytes accumulate and transmit damaged mitochondria due to an inability to activate mitophagy. Dev Biol 426(1):126–138. https://doi.org/10.1016/j.ydbio.2017.04.005
Wu LL, Russell DL, Wong SL et al (2015) Mitochondrial dysfunction in oocytes of obese mothers: transmission to offspring and reversal by pharmacological endoplasmic reticulum stress inhibitors. Development 142(4):681–691. https://doi.org/10.1242/dev.114850
Park JH, Stoffers DA, Nicholls RD, Simmons RA (2008) Development of type 2 diabetes following intrauterine growth retardation in rats is associated with progressive epigenetic silencing of Pdx1. J Clin Invest 118(6):2316–2324. https://doi.org/10.1172/JCI33655
Sandovici I, Smith NH, Nitert MD et al (2011) Maternal diet and aging alter the epigenetic control of a promoter-enhancer interaction at the Hnf4a gene in rat pancreatic islets. Proc Natl Acad Sci U S A 108(13):5449–5454. https://doi.org/10.1073/pnas.1019007108
Fernandez-Twinn DS, Wayman A, Ekizoglou S, Martin MS, Hales CN, Ozanne SE (2005) Maternal protein restriction leads to hyperinsulinemia and reduced insulin-signaling protein expression in 21-mo-old female rat offspring. Am J Physiol Regul Integr Comp Physiol 288(2):R368–R373. https://doi.org/10.1152/ajpregu.00206.2004
Ding GL, Wang FF, Shu J et al (2012) Transgenerational glucose intolerance with Igf2/H19 epigenetic alterations in mouse islet induced by intrauterine hyperglycemia. Diabetes 61(5):1133–1142. https://doi.org/10.2337/db11-1314
Berends LM, Fernandez-Twinn DS, Martin-Gronert MS, Cripps RL, Ozanne SE (2013) Catch-up growth following intra-uterine growth-restriction programmes an insulin-resistant phenotype in adipose tissue. Int J Obes 37(8):1051–1057. https://doi.org/10.1038/ijo.2012.196
Fernandez-Twinn DS, Alfaradhi MZ, Martin-Gronert MS et al (2014) Downregulation of IRS-1 in adipose tissue of offspring of obese mice is programmed cell-autonomously through post-transcriptional mechanisms. Molecular Metabolism 3(3):325–333. https://doi.org/10.1016/j.molmet.2014.01.007
Alfaradhi MZ, Kusinski LC, Fernandez-Twinn DS et al (2016) Maternal obesity in pregnancy developmentally programs adipose tissue inflammation in young, lean male mice offspring. Endocrinology 157(11):4246–4256. https://doi.org/10.1210/en.2016-1314
de Almeida FJ, Duque-Guimarães D, Carpenter AA, Loche E, Ozanne SE (2017) A post-weaning obesogenic diet exacerbates the detrimental effects of maternal obesity on offspring insulin signaling in adipose tissue. Sci Rep 7(1):44949. https://doi.org/10.1038/srep44949
Butruille L, Marousez L, Pourpe C et al (2019) Maternal high-fat diet during suckling programs visceral adiposity and epigenetic regulation of adipose tissue stearoyl-CoA desaturase-1 in offspring. Int J Obes. https://doi.org/10.1038/s41366-018-0310-z
Ferland-McCollough D, Fernandez-Twinn DS, Cannell IG et al (2012) Programming of adipose tissue miR-483-3p and GDF-3 expression by maternal diet in type 2 diabetes. Cell Death Differ 19(6):1003–1012. https://doi.org/10.1038/cdd.2011.183
Marco A, Kisliouk T, Tabachnik T, Meiri N, Weller A (2014) Overweight and CpG methylation of the Pomc promoter in offspring of high-fat-diet-fed dams are not “reprogrammed” by regular chow diet in rats. FASEB J 28(9):4148–4157. https://doi.org/10.1096/fj.14-255620
Vucetic Z, Kimmel J, Totoki K, Hollenbeck E, Reyes TM (2010) Maternal high-fat diet alters methylation and gene expression of dopamine and opioid-related genes. Endocrinology 151(10):4756–4764. https://doi.org/10.1210/en.2010-0505
Stevens A, Begum G, White A (2011) Epigenetic changes in the hypothalamic pro-opiomelanocortin gene: a mechanism linking maternal undernutrition to obesity in the offspring? Eur J Pharmacol 660(1):194–201. https://doi.org/10.1016/j.ejphar.2010.10.111
Begum G, Stevens A, Smith EB et al (2012) Epigenetic changes in fetal hypothalamic energy regulating pathways are associated with maternal undernutrition and twinning. FASEB J 26(4):1694–1703. https://doi.org/10.1096/fj.11-198762
Berends LM, Dearden L, Tung YCL, Voshol P, Fernandez-Twinn DS, Ozanne SE (2018) Programming of central and peripheral insulin resistance by low birthweight and postnatal catch-up growth in male mice. Diabetologia 61(10):2225–2234. https://doi.org/10.1007/s00125-018-4694-z
Chiang PK, Gordon RK, Tal J et al (1996) S-adenosylmethionine and methylation. FASEB J 10(4):471–480. https://doi.org/10.1096/fasebj.10.4.8647346
Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM (1998) A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 92(6):829–839. https://doi.org/10.1016/S0092-8674(00)81410-5
Patti ME, Butte AJ, Crunkhorn S et al (2003) Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: potential role of PGC1 and NRF1. Proc Natl Acad Sci U S A 100(14):8466–8471. https://doi.org/10.1073/pnas.1032913100
Brøns C, Jacobsen S, Nilsson E et al (2010) Deoxyribonucleic acid methylation and gene expression of PPARGC1A in human muscle is influenced by high-fat overfeeding in a birth-weight-dependent manner. J Clin Endocrinol Metab 95(6):3048–3056. https://doi.org/10.1210/jc.2009-2413
Gillberg L, Jacobsen SC, Rönn T, Brøns C, Vaag A (2014) PPARGC1A DNA methylation in subcutaneous adipose tissue in low birth weight subjects — impact of 5 days of high-fat overfeeding. Metabolism 63(2):263–271. https://doi.org/10.1016/j.metabol.2013.10.003
Hjort L, Jørgensen SW, Gillberg L et al (2017) 36 h fasting of young men influences adipose tissue DNA methylation of LEP and ADIPOQ in a birth weight-dependent manner. Clin Epigenetics 9(1):40. https://doi.org/10.1186/s13148-017-0340-8
Rönn T, Volkov P, Davegårdh C et al (2013) A six months exercise intervention influences the genome-wide DNA methylation pattern in human adipose tissue. PLoS Genet 9(6):e1003572. https://doi.org/10.1371/journal.pgen.1003572
Alibegovic AC, Sonne MP, Højbjerre L et al (2010) Insulin resistance induced by physical inactivity is associated with multiple transcriptional changes in skeletal muscle in young men. Am J Physiol Endocrinol Metab 299(5):E752–E763. https://doi.org/10.1152/ajpendo.00590.2009
McGee SL, Fairlie E, Garnham AP, Hargreaves M (2009) Exercise-induced histone modifications in human skeletal muscle. J Physiol 587(24):5951–5958. https://doi.org/10.1113/jphysiol.2009.181065
Fernandez-Twinn DS, Gascoin G, Musial B et al (2017) Exercise rescues obese mothers’ insulin sensitivity, placental hypoxia and male offspring insulin sensitivity. Sci Rep 7(1):44650. https://doi.org/10.1038/srep44650
Stanford KI, Lee MY, Getchell KM, So K, Hirshman MF, Goodyear LJ (2015) Exercise before and during pregnancy prevents the deleterious effects of maternal high-fat feeding on metabolic health of male offspring. Diabetes 64(2):427–433. https://doi.org/10.2337/db13-1848
Stanford KI, Takahashi H, So K et al (2017) Maternal exercise improves glucose tolerance in female offspring. Diabetes 66(8):2124–2136. https://doi.org/10.2337/db17-0098
Smallwood SA, Kelsey G (2012) De novo DNA methylation: a germ cell perspective. Trends Genet 28(1):33–42. https://doi.org/10.1016/j.tig.2011.09.004
Funding
DSF-T and SEO are supported by MRC-MDU programme grants (MC_UU_12012/4 and and MC_UU_00014/4)) and by a British Heart Foundation Programme grant (RG/17/12/33167). LH was funded by the Danish Diabetes Academy, funded by the Novo Nordisk Foundation. BN is funded by an NHMRC (Australia) CJ Martin Fellowship (#1072966) and New Investigator Grant (#1157556).
Author information
Authors and Affiliations
Contributions
All authors were responsible for drafting the article and revising it critically for important intellectual content. All authors approved the version to be published.
Corresponding authors
Ethics declarations
The authors declare that there is no duality of interest associated with this manuscript.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Denise S. Fernandez-Twinn, Line Hjort and Boris Novakovic are joint first authors. Susan E. Ozanne and Richard Saffery are joint senior authors.
Electronic supplementary material
Slideset of figures
(PPTX 343 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Fernandez-Twinn, D.S., Hjort, L., Novakovic, B. et al. Intrauterine programming of obesity and type 2 diabetes. Diabetologia 62, 1789–1801 (2019). https://doi.org/10.1007/s00125-019-4951-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-019-4951-9