Abstract
Aims/hypothesis
The euglycaemic–hyperinsulinaemic clamp is the gold-standard method for measuring insulin sensitivity, but is less suitable for large clinical trials. Thus, several indices have been developed for evaluating insulin sensitivity from the oral glucose tolerance test (OGTT). However, most of them yield values different from those obtained by the clamp method. The aim of this study was to develop a new index to predict clamp-derived insulin sensitivity (M value) from the OGTT-derived oral glucose insulin sensitivity index (OGIS).
Methods
We analysed datasets of people that underwent both a clamp and an OGTT or meal test, thereby allowing calculation of both the M value and OGIS. The population was divided into a training and a validation cohort (n = 359 and n = 154, respectively). After a stepwise selection approach, the best model for M value prediction was applied to the validation cohort. This cohort was also divided into subgroups according to glucose tolerance, obesity category and age.
Results
The new index, called PREDIcted M (PREDIM), was based on OGIS, BMI, 2 h glucose during OGTT and fasting insulin. Bland–Altman analysis revealed a good relationship between the M value and PREDIM in the validation dataset (only 9 of 154 observations outside limits of agreement). Also, no significant differences were found between the M value and PREDIM (equivalence test: p < 0.0063). Subgroup stratification showed that measured M value and PREDIM have a similar ability to detect intergroup differences (p < 0.02, both M value and PREDIM).
Conclusions/interpretation
The new index PREDIM provides excellent prediction of M values from OGTT or meal data, thereby allowing comparison of insulin sensitivity between studies using different tests.
Similar content being viewed by others

Introduction
Insulin resistance contributes to the deterioration of postprandial glucose homeostasis, thereby playing a fundamental role in the pathophysiology of type 2 diabetes [1]. Insulin resistance is also a common feature of obesity, non-alcoholic fatty liver disease and cardiovascular diseases [2, 3].
The reference method to quantify insulin sensitivity is the euglycaemic–hyperinsulinaemic clamp technique [4]. The clamp-derived M value, based on steady-state glucose infusion rates (GIR), yields a measure of insulin sensitivity under conditions of constant glycaemia and hyperinsulinaemia. Despite its accuracy and precision, the procedure is time-consuming and costly, and its use for screening or larger clinical studies is therefore limited. The (frequently sampled) intravenous glucose tolerance test, coupled with modelling analysis, also provides an insulin sensitivity index, which has been thoroughly validated against the clamp. However, it shares several limitations with the clamp, despite recent methodological advances [5].
The OGTT was originally employed to classify glucose tolerance and to diagnose diabetes but, more recently, it has also been used to evaluate beta cell function and insulin secretion, as well as insulin sensitivity [6]. Several OGTT-based indices typically show acceptable correlation with the clamp-derived M value. Nonetheless, these OGTT indices have different measurement units than the M value, thus making direct comparisons difficult. This prompted the present study, which aimed at developing and validating a method to predict clamp-derived insulin sensitivity from an OGTT-derived index. Among several indices, as the starting point for the prediction of the M value we selected the oral glucose insulin sensitivity index (OGIS) [7], which has the advantage of representing glucose clearance. Thus, OGIS has a specific physiological basis, compared with other totally empirical indices. Also, in a previous study comparing some OGTT-derived indices, OGIS performed better than the other indices analysed [8]. In our approach, the clamp index is predicted from OGIS in combination with easily obtained measurements, such as anthropometric or other simply assessed variables.
Methods
Participants and experimental procedures
This study analyses data collected in previous studies, each carried out in agreement with the Declaration of Helsinki and upon approval by the respective local ethics committees [9,10,11,12,13,14]. Participants were studied at (1) the German Diabetes Center, Düsseldorf, Germany; (2) the Karl-Landsteiner Institute for Endocrinology and Metabolism and 1st Medical Department, Hanusch Hospital, Vienna, Austria, and Department of Internal Medicine III, Medical University of Vienna, Vienna, Austria; (3) the Steno Diabetes Center, Gentofte, Denmark; (4) the Institute of Endocrinology, Prague, Czech Republic; and (5) the Clinical Research Center of the University of Texas Health Science Center, San Antonio, TX, USA.
Participants who attended the German Diabetes Center did not have type 2 diabetes at the time of recruitment, though some had a positive family history of type 2 diabetes [9]. Participants recruited in Vienna either had normal glucose tolerance (NGT), impaired glucose tolerance (IGT) or type 2 diabetes [10]. Participants from the Steno Diabetes Center were recruited from the Inter99 study and had either NGT, isolated IGT, or isolated impaired fasting glucose (IFG) [11]. Participants from the Prague Institute of Endocrinology were part of two datasets: the first one included bariatric patients with type 2 diabetes [12], the second included patients with conditions known to affect insulin sensitivity (obesity and/or polycystic ovary syndrome), without known type 2 diabetes [13]. Participants from the San Antonio Clinical Research Center were recruited through advertising within the medical centre and in local newspapers, and had glucose tolerance spanning from normal to type 2 diabetes [14]. Experimental clamp procedures at the five centres have been reported in detail in the original studies [9,10,11,12,13,14]. For all participants, the M value was calculated as the glucose infusion rate during the last 20–30 min of the test with space correction when appropriate [4]. Participants from Germany/Austria, Denmark, Texas, and from the second Czech Republic dataset also underwent a 2–3 h 75 g OGTT, while those from the first Czech Republic dataset received a standardised liquid mixed-meal test (MMT). Values from either test, the OGTT or MMT, depending on which were available, were used for the calculation of the OGIS index, as a previous study demonstrated the equivalence of OGIS from OGTT and MMT [15].
Calculation of OGIS
The approach for the derivation of OGIS has previously been provided in full [7]. Here, we briefly present its formula for the 3 h OGTT or MTT test (with glucose and insulin in SI units):
where G0, G120, G180 is glucose at 0, 120, 180 min, I0 and I120 is insulin at 0 and 120 min, p1 = 2.89, p2 = 1618, p3 = 779, p4 = 2642, p5 = 11.5 × 10−3, p6 = 117, V = 104 (glucose distribution volume, ml/m2), T = 60 (time interval between G180 and G120, min), Gcl = 5 (typical clamp glucose concentration, mmol/l), and D0 is glucose dose of the OGTT or MTT (in mmol/m2, i.e. normalised for body surface area); sqrt is the square root operator. In the case of the 2 h test, the OGIS formula is similar, but G180 is replaced by G120, G120 and I120 are replaced by G90 and I90, respectively, and T = 30; p1–p6 parameters become: p1 = 6.50, p2 = 1951, p3 = 4514, p4 = 792, p5 = 11.8 × 10−3, p6 = 173.
Prediction model of clamp-derived insulin sensitivity
The prediction of the M value from OGIS was based on the development of a multivariable model. We hypothesised that when some individuals are studied with an oral glucose challenge, data on some basic variables are always available: sex, age, BMI, fasting glucose and insulin concentrations and 2 h glucose concentration. These variables were considered as possible predictors of the M value along with OGIS. From the above variables, we also derived some categorical variables: glucose tolerance status (according to ADA 2010 criteria, i.e. NGT, IFG and/or IGT, and type 2 diabetes [16]), the obesity category (BMI < 25 kg/m2, lean; BMI ≥ 30 kg/m2, obese; overweight otherwise) and age category (elderly if ≥ 50 years). The total dataset (513 individuals) was randomly split into training and validation datasets (70% and 30%, respectively, according to common practice [17]). In the training dataset, all potential predictors were included in a linear regression model providing R2 statistics. We then applied the stepwise model selection approach, based on Akaike’s information criterion (AIC), to determine the optimal prediction model (the lower the AIC value, the better the model) [18], including both backward and forward search strategies. The optimal prediction model thus identified was then applied to the validation dataset.
Insulin sensitivity in subgroups
In the validation dataset, participants were divided into subgroups according to the following categories: glucose tolerance, obesity and age. We then analysed possible differences in insulin sensitivity among the subgroups, according to both the real (clamp–derived) and the model-predicted M value.
Statistics
All analyses indicated in the Methods were performed in R (The R Foundation for Statistical Computing Platform, Vienna, Austria). Data are presented as mean ± standard error (SEM), unless otherwise specified. In both the training and validation datasets, comparison between observed and predicted M value was performed by linear regression and test of equivalence [19] (the two one-sided paired t test) on logarithmically transformed values, in order to achieve homoscedastic prediction models. Bland–Altman plots including limits of agreement were also reported. Further validation of M value prediction was performed by leave-one-out cross-validation (LOOCV) on the training dataset, and related cross-validated R2 statistics [20]. In the validation dataset, tests on subgroups were performed using ANOVA (on logarithmically transformed data). A two-sided p value < 0.05 was considered statistically significant.
Results
Model development
Table 1 presents the variables tested for M value prediction in both the training and the validation datasets. All the variables selected for M value prediction (i.e. OGIS, sex, age, BMI, fasting glucose and insulin and 2 h glucose) were included in the initial multivariable regression model. However, the majority of them were not significant in the overall model, which is likely to be due to the excessive number of variables. Using the stepwise approach, we then searched for the optimal model, which identified OGIS, BMI, 2 h glucose (2hGLU) and fasting insulin (INSf) as the variables to be included (AIC = −712.73 vs AIC = −591.28 for the model including OGIS alone). The new index for M value prediction, called PREDIcted M (PREDIM), was calculated as follows:
where A = 2.8846219, B = 0.5208520, C = −0.8223363, D = −0.4191242, E = −0.2427896.
Comparison with M values
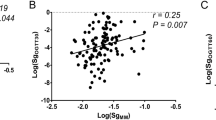
Based on eq. (2) we calculated the PREDIM values in the training dataset, and compared them with the corresponding real M values. Linear regression analysis yielded an adjusted R2 of 0.733, p < 0.0001 (Fig. 1a). Of note, when predicting the M value with OGIS alone, the R2 value was lower (adjusted R2 = 0.619, p < 0.0001). The estimated variance of the error in eq. (2) was 0.135. The Bland–Altman plot showed that only 18 out of 359 observations were outside the limits of agreement (Fig. 2a), though it should be acknowledged that the plot suggests a small downward trend, this meaning somehow lower agreement for the extreme values (very low or very high values). According to the equivalence test, the M value and PREDIM value were virtually identical (mean of the difference between log e -transformed M value and PREDIM value was −5.95 × 10−16, with p < 0.0001 when the equivalence bound was set to 0.1). This means having tested that the PREDIM values in the original, not log e -transformed units, are within 10.5% of the M values (e0.1 is in fact equal to 1.105). Notably, the test remained significant with the equivalence bound lowered to 0.032, i.e. equivalence in the original units within 3.2%.
Linear regression plot in the training dataset (a) and in the validation dataset (b). Regression equations (solid lines) are y = 1.00x – 0.01, R2 = 0.733, p < 0.0001 (a), and y = 1.02x + 0.01, R2 = 0. 692, p < 0.0001 (b). 95% CIs and prediction intervals are also reported (dashed lines and solid thick lines, respectively)
Model validation
When applying eq. (2) to the validation dataset, we found a tight relationship between PREDIM values and real M values, i.e. adjusted R2 = 0.692, p < 0.0001 by linear regression (Fig. 1b), which was still superior to the variance explained by OGIS alone (adjusted R2 = 0.587). The estimated variance of the error in eq. (2) was 0.151. Again, according to the equivalence test, the M value and PREDIM value were very similar, showing a mean of the difference equal to 0.021, p < 0.0063 with an equivalence bound of 0.1 (the test remained significant when the equivalence bound was lowered, to 0.073). The corresponding Bland–Altman plot detected only nine of 154 observations outside the limits of agreement (Fig. 2b), though again suggesting a small downward trend. For further validation, we used the LOOCV method, which yielded a cross-validated R2 of 0.728, in agreement with the linear regression results reported above.
Subgroup discrimination
In the validation dataset, when participants were divided into subgroups, we found complete agreement between the M value and PREDIM value, which were both significantly decreased from NGT to type 2 diabetes, as expected (p < 0.0001 for both the real M value and PREDIM value; Fig. 3a,b). Similarly, both the M and PREDIM values showed significantly decreased levels across obesity (p < 0.0001 for both; Fig. 3c,d), and from young to elderly (p < 0.02 for the M value, p < 0.001 for the PREDIM value; Fig. 3e,f). In addition, we have performed the equivalence test in the single subgroups. For subgroups based on glucose tolerance, the M value and PREDIM value were found to be similar, down to an equivalence bound of 0.13 for NGT and 0.11 for both impaired glucose regulation (IFG and/or IGT) and type 2 diabetes participants. For subgroups based on BMI, the lowest equivalence bounds were 0.18 for lean, 0.16 for overweight and 0.10 for obese participants, whereas for subgroups based on age, the lowest equivalence bounds were 0.08 for young and 0.12 for elderly participants.
Bar chart (mean ± SEM) for observed M (left) and predicted M (right) in the validation dataset for participants stratified by glucose tolerance (a, b), degree of obesity (c, d) and age (e, f). IGR, impaired glucose regulation (IFG and/or IGT); T2DM, type 2 diabetes. The number of individuals in each subgroup is reported in Table 1
Discussion
This report presents a new method to predict clamp-derived whole body insulin sensitivity (M value) from an OGTT or MMT performed in a large cohort of individuals with varying degrees of glucose tolerance. We developed an index, called PREDIM, calculated from OGIS [7] and other simply assessed variables. The reliability of the proposed method was demonstrated in an independent sub-sample of participants. This method yielded excellent agreement between the real (clamp-derived) and the predicted M value, both in terms of mutual relationships and in terms of the ability to detect differences among subgroups of participants.
The relevance of this study lies in the ability to directly compare OGTT-based and clamp-based insulin sensitivity values obtained in the same individual within a short period of time. This will be of special interest for the comparison of data from different studies, where participants have been examined by one test or the other. Specifically, the new formula makes it possible to calculate the M value for large-scale studies where only OGTT or MMT data are available, and to compare them with clamp-derived M values obtained in small experimental studies with intensive phenotyping.
Among all the available methods for the estimation of insulin sensitivity from an OGTT or MMT, we opted for OGIS because previous studies have proven its superior performance compared with other indices [8]. Moreover, OGIS is based on a more solid physiological basis, since it describes the glucose clearance, while most other indices are purely empirical. Another advantage of OGIS resides in its applicability to both OGTT and any kind of meal test, only requiring the input of the appropriate amount of administered glucose into the calculation [15].
The new formula for M value prediction features OGIS plus other predictors. The selected additional predictors are simple and always available when performing an OGTT. In particular, BMI, 2 h glucose and fasting insulin together improved M value prediction. For the development of the prediction model, it was first necessary to select two datasets: the development (training) dataset and the validation dataset. Here, we randomly split the total dataset into the training and validation sets, in percentages of 70% and 30%, respectively. To our knowledge, there are no precise recommendations about percentages for training and validation, thus we chose the commonly used split of 70% and 30% [17]. This choice yielded a heterogeneous training dataset that included male and female individuals covering a wide range of ages, BMIs and glucose tolerance (which ranged from NGT to severe type 2 diabetes, and, as expected, insulin sensitivity varied manifold between subgroups). In the validation dataset, the relationship between predicted and observed M values was remarkably good, as assed by linear regression analysis, equivalence statistics, and Bland−Altman plot.
As prediction models may also include non-linear terms, we carried out several supplementary analyses. However, we did not find any substantial improvement (not shown); therefore, we concluded that the proposed linear multivariable model, i.e. equation (2), represents the easiest and, at the same time, the most appropriate solution.
In this study, the number of participants with a long duration of type 2 diabetes was relatively small. This may mean that severe insulin resistance is under-represented in the database. However, there was a wide range of M values in the training dataset (ranging from 0.5 to 15.2 mg kg−1 min−1). One previous study suggested an M value of 4.9 mg kg−1 min−1 as a cut-off level for insulin resistance [21]. Since in our training dataset 67% of M values were lower than this cut-off level (with the lowest values being about 10% of the cut-off), we claim that our formula will perform satisfactorily in individuals with severe insulin resistance.
In conclusion, we have exploited rigorous statistical techniques to develop an index (PREDIM) that predicts clamp insulin sensitivity (M value) from an OGIS value in combination with some basic variables easily available. The steps that need to be performed for M value prediction are: (1) use of eq. (2) to get PREDIM in log units, and (2) reverse log transformation [PREDIM = e log(PREDIM)] to obtain PREDIM in the traditional M units (mg kg−1 min−1). We have found that the method provides excellent prediction of the real M value and allows the comparison of insulin sensitivity from different investigations and different groups of participants, who may have been studied with different tests.
Data availability
The data are available on request from the authors.
Abbreviations
- AIC:
-
Akaike’s information criterion
- GIR:
-
Glucose infusion rates
- IFG:
-
Impaired fasting glucose
- IGT:
-
Impaired glucose tolerance
- LOOCV:
-
Leave-one-out cross-validation
- NGT:
-
Normal glucose tolerance
- MMT:
-
Mixed-meal test
- OGIS:
-
Oral glucose insulin sensitivity index
- PREDIM:
-
PREDIcted M
References
DeFronzo RA (2009) Banting lecture. From the triumvirate to the ominous octet: a new paradigm for the treatment of type 2 diabetes. Diabetes 58:773–795
Szendroedi J, Phielix E, Roden M (2011) The role of mitochondria in insulin resistance and type 2 diabetes mellitus. Nat Rev Endocrinol 13:92–103
Ferrannini E (2006) Is insulin resistance the cause of the metabolic syndrome? Ann Med 38:42–51
DeFronzo RA, Tobin JD, Andres R (1979) Glucose clamp technique: a method for quantifying insulin secretion and resistance. Am J Phys 237:E214–E223
Tura A, Sbrignadello S, Succurro E, Groop L, Sesti G, Pacini G (2010) An empirical index of insulin sensitivity from short IVGTT: validation against the minimal model and glucose clamp indices in patients with different clinical characteristics. Diabetologia 53:144–152 Erratum in Diabetologia 2010;53:1245
Pacini G, Mari A (2003) Methods for clinical assessment of insulin sensitivity and beta cell function. Best Pract Res Clin Endocrinol Metab 17:305–322
Mari A, Pacini G, Murphy E, Ludvik B, Nolan JJ (2001) A model-based method for assessing insulin sensitivity from the oral glucose tolerance test. Diabetes Care 24:539–548
Mari A, Pacini G, Brazzale AR, Ahrén B (2005) Comparative evaluation of simple insulin sensitivity methods based on the oral glucose tolerance test. Diabetologia 48:748–751
Szendroedi J, Yoshimura T, Phielix E et al (2014) Role of diacylglycerol activation of PKCθ in lipid-induced muscle insulin resistance in humans. Proc Natl Acad Sci U S A 111:9597–9602
Szendroedi J, Kaul K, Kloock L et al (2014) Lower fasting muscle mitochondrial activity relates to hepatic steatosis in humans. Diabetes Care 37:468–474
Faerch K, Vaag A, Holst JJ, Glümer C, Pedersen O, Borch-Johnsen K (2008) Impaired fasting glycaemia vs impaired glucose tolerance: similar impairment of pancreatic alpha and beta cell function but differential roles of incretin hormones and insulin action. Diabetologia 51:853–861
Bradnova O, Kyrou I, Hainer V et al (2014) Laparoscopic greater curvature plication in morbidly obese women with type 2 diabetes: effects on glucose homeostasis, postprandial triglyceridemia and selected gut hormones. Obes Surg 24:718–726
Vrbíková J, Cibula D, Dvoráková K et al (2004) Insulin sensitivity in women with polycystic ovary syndrome. J Clin Endocrinol Metab 89:2942–2945
Ferrannini E, Gastaldelli A, Miyazaki Y, Matsuda M, Mari A, DeFronzo RA (2005) β-cell function in subjects spanning the range from normal glucose tolerance to overt diabetes: a new analysis. J Clin Endocrinol Metab 90:493–500
Mari A, Gastaldelli A, Foley JE, Pratley RE, Ferrannini E (2005) Beta-cell function in mild type 2 diabetic patients: effects of 6-month glucose lowering with nateglinide. Diabetes Care 28:1132–1138
American Diabetes Association (2010) Diagnosis and classification of diabetes mellitus. Diabetes Care 33(Suppl 1):S62–S629
Crowther PS, Cox RJ (2005) A method for optimal division of data sets for use in neural networks. In: Khosla R, Howlett RJ, Jain LC (eds) Knowledge-based intelligent information and engineering systems. KES 2005. Lecture notes in computer science, vol. 3684, Springer, Berlin, Heidelberg, pp 1–7
Hocking RR (1976) The analysis and selection of variables in linear regression. Biometrics 32:1–49
Robinson AP, Froese RE (2004) Model validation using equivalence tests. Ecol Model 176:349–358
Michiels S, Le Maitre A, Buyse M et al (2009) Surrogate endpoints for overall survival in locally advanced head and neck cancer: meta-analyses of individual patient data. Lancet Oncol 10:341–350
Tam CS, Xie W, Johnson WD, Cefalu WT, Redman LM, Ravussin E (2012) Defining insulin resistance from hyperinsulinemic-euglycemic clamps. Diabetes Care 35:1605–1610
Acknowledgements
The authors wish to thank G. Kacerovsky-Bielesz and A. Brehm (Hanusch-Krankenhaus, Vienna, Austria) and A. Schmid, M. Chmelik and M. Fritsch (Medical University of Vienna, Vienna, Austria) for their help in clarifying some aspects of the data.
Contribution statement
AT analysed the data, developed the model, and wrote the manuscript; GC and CG contributed to the analyses of the data and model development, and drafted the manuscript; JS, KF, JV, EF collected or contributed to collect the data, contributed to the interpretation of the results, and drafted the manuscript; GP contributed to the design of the study, to data analyses and results interpretation, and revised the manuscript critically; MR designed the study, contributed to data analyses and results interpretation, and revised the manuscript critically. All authors approved the final version. MR is the guarantor of this work.
Funding
This study was supported in part by the Ministry of Science and Research of the State of North Rhine-Westphalia (MIWF NRW) and the German Federal Ministry of Health (BMG) to the German Diabetes Center (DDZ), by a grant from the Federal Ministry for Research (BMBF) to the German Center for Diabetes Research (DZD e.V.) as well as by grants from the Helmholtz Alliance to Universities (ICEMED), the German Research Foundation (DFG, SFB 1116), German Diabetes Association (DDG) and the Schmutzler-Stiftung. Data on participants recruited in Vienna were analysed in studies supported by the European Foundation for the Study of Diabetes (Novo Nordisk type 2 diabetes grant, GSK grant), the Austrian Science Foundation (P15656), and the Austrian National Bank (OENB 11459) to MR, and by a Research Grant Award by the Austrian Diabetes Association to Gertrud Kacerovsky-Bielesz, Hanusch-Krankenhaus, Vienna, Austria. The Copenhagen study was supported by the Danish Ministry of Science, Technology and Innovation, the Danish Diabetes Association, the Novo Nordisk Foundation, the Foundation of Gerda and Aage Haensch, and by an EXGENESIS grant (005272) from the European Union. The Prague studies were supported by the grant of the European Foundation for the Study of Diabetes (EFSD) and by the Ministry of Health, Czech Republic - conceptual development of research organization (Institute of Endocrinology – EU 00023761). The San Antonio Metabolism Study was supported by funds from the Italian Ministry of University and Scientific Research (2001065883-001).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that there is no duality of interest associated with this manuscript.
Rights and permissions
About this article
Cite this article
Tura, A., Chemello, G., Szendroedi, J. et al. Prediction of clamp-derived insulin sensitivity from the oral glucose insulin sensitivity index. Diabetologia 61, 1135–1141 (2018). https://doi.org/10.1007/s00125-018-4568-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-018-4568-4