Abstract
Aim/hypothesis
Abnormal activation of hepatic gluconeogenesis leads to hyperglycaemia. However, the molecular mechanisms underlying dysregulated hepatic gluconeogenesis remain to be fully defined. Here, we explored the physiological role of Krüppel-like factor 10 (KLF10) in regulating hepatic glucose metabolism in mice.
Methods
Hepatic KLF10 expression in wild-type C57BL/6J mice, the db/db mouse model of diabetes, the ob/ob mouse model of obesity and high-fat-diet-induced obese (DIO) mice was measured. Adenoviruses expressing Klf10 or Klf10-specific short-hairpin RNA were injected into wild-type C57BL/6J mice, db/db or DIO mice. Expression of gluconeogenic genes in the liver and blood glucose levels were measured. GTTs and pyruvate tolerance tests were performed. The molecular mechanism by which KLF10 regulates hepatic glucose metabolism was explored.
Results
Hepatic KLF10 expression was regulated by nutritional status in wild-type mice and upregulated in diabetic, obese and DIO mice. Overexpression of KLF10 in primary hepatocytes increased the expression of gluconeogenic genes and cellular glucose output. C57BL/6J mice with KLF10 overexpression in the liver displayed increased blood glucose levels and impaired glucose tolerance. Conversely, hepatic KLF10 knockdown in db/db and DIO mice decreased blood glucose levels and improved glucose tolerance. Furthermore, luciferase reporter gene assay and chromatin immunoprecipitation analysis indicated that KLF10 activates Pgc-1α (also known as Ppargc1a) gene transcription via directly binding to its promoter region.
Conclusions/interpretation
KLF10 is an important regulator of hepatic glucose metabolism and modulation of KLF10 expression in the liver may be an attractive approach for the treatment of type 2 diabetes.
Similar content being viewed by others
Introduction
In mammals, blood glucose levels are maintained within a relatively narrow range through the regulation of glucose production by the liver and glucose uptake by peripheral tissues. Maintenance of blood glucose homeostasis protects the body against hyperglycaemia after ingestion of a carbohydrate-rich meal and against hypoglycaemia during fasting [1, 2]. Under fasting conditions, circulating glucagon and glucocorticoid levels increase to stimulate the expression of gluconeogenic genes, which increase glucose secretion by the liver [3]. However, the abnormal activation of hepatic gluconeogenesis contributes to hyperglycaemia in diabetes [4]. Therefore, efforts to uncover the molecular mechanisms that regulate hepatic gluconeogenesis are crucial to the development of new therapeutic strategies for the treatment of diabetes [1].
Peroxisome proliferator-activated receptor, gamma, coactivator 1α (PGC-1α) is a transcriptional coactivator that plays a central role in the regulation of cellular energy metabolism. Hepatic expression of PGC-1α is elevated in type 1 and type 2 diabetes mellitus [1, 5, 6]. In the fasted state, PGC-1α expression is induced by increased circulating glucagon and catecholamines via the cAMP and cAMP response element-binding protein (CREB)/transducer of regulated CREB activity (TORC) pathways [5, 7]. In turn, PGC-1α stimulates the expression of Pck1 and G6pc, two key gluconeogenic genes encoding PEPCK and glucose-6-phosphatase (G6Pase), respectively, through directly interacting with and coactivating several transcription factors such as hepatic nuclear factor-4 (HNF4), glucocorticoid receptor (GR), and forkhead box O1 (FOXO1) [1, 8, 9]. In contrast, insulin/Akt suppresses PGC-1α activity under feeding condition [10,11,12,13].
Krüppel-like factor 10 (KLF10), which was originally named TGF-β inducible early gene-1 (TIEG1), is a member of Krüppel-like family of transcription factors (KLFs). The KLFs belong to a subclass of Cys2/His2 zinc-finger DNA-binding proteins and are critical regulators of cellular homeostasis, with roles in growth, development and programmed cell death [14, 15]. KLF10 plays important roles in TGF-β-mediated cell growth, apoptosis and differentiation [16]. KLF10 is ubiquitously expressed, with high expression levels in the liver, and its gene variants may correlate with the development of type 2 diabetes [17,18,19]. Recently, KLF10 has been reported to be associated with non-alcoholic steatohepatitis (NASH) [20].
In the present study, we investigated the role of KLF10 in the regulation of hepatic glucose metabolism.
Methods
Animals and experimental design
Male db/db, db/m (heterozygotes for leptin receptor-deficient mice, littermate control mice of db/db), ob/ob and C57BL/6J mice aged 6–8 weeks were purchased from the Model Animal Research Center of Nanjing University (Nanjing, China) and housed and maintained in 12 h light/dark photoperiod with unrestricted water and food. In order to establish a DIO model and a normal diet control model, wild-type C57BL/6J mice were fed either ad libitum or a high-fat diet (45% fat; Research Diets, New Brunswick, NJ, USA) with free access to water. All animal experiments were carried out under protocols approved by the Animal Research Committee of the Institute of Laboratory Animals, Chinese Academy of Medical Sciences and Peking Union Medical College. For fasting experiments, food was removed for 6 h before the mice were killed. Mice were injected i.v. through the tail vein with adenovirus expressing green fluorescent protein (Ad-gfp), adenovirus expressing KLF10 (Ad-Klf10), control adenovirus expressing short-hairpin (sh)RNA against luciferase (Ad-shCtrl) or adenovirus expressing shRNA against KLF10 (Ad-shKlf10) (0.5–1.5 × 109 active viral particles in 200 μl saline [54 mmol/l NaCl]). At 7–9 days after infection, mice were fasted for 6 h and then killed. Their livers and plasma were collected for further analysis. For animal experiments, all mice were grouped randomly, and experimenters were blind to group assignment and outcome assessment. No data, samples or animals were excluded.
Preparation of recombinant adenoviruses and expression plasmids
The full-length mouse Klf10 gene was amplified by PCR from C57BL/6J mouse liver cDNA, and myc-tagged Klf10 was cloned into pcDNA3.1 (Invitrogen, Carlsbad, CA, USA) using the following PCR primer pairs: 5′-CGCCGGTACCATGCTCAACTTCGGCGCT TCTCTCC-3′ (forward) and 5′-CGCGCTCGAGTCACAGATCCTCTTCTGAGATGAGTTTTTGTTCCTGTGCGGAAGCAGGGGT-3′ (reverse). Adenovirus expressing vectors were purchased from Promega (Madison, WI, USA). Recombinant Ad-Klf10 was generated as previously described [21]. The mouse Pgc-1α gene promoter (−2525 to +82 bp) was amplified by PCR using mouse genomic DNA and inserted into a pGL3-basic luciferase reporter vector (−2525Luc). A series of 5′ truncated constructs of the Pgc-1α gene promoter (−729Luc, −171Luc, −34Luc) were prepared by PCR using −2525Luc as a template. The primers are shown in electronic supplementary material (ESM) Table 1.
RNA interference
shRNA-encoding DNA sequences were synthesised by Invitrogen and constructed into adenovirus plasmids (pAdTrack-U6 vectors), and adenoviruses were generated according to previously described procedures [21]. The sequence of small interfering (si)RNA against luciferase (siControl) was 5′-CTTACGCTGAGTACTTCGA-3′, and the sequence of siRNA against Klf10 (siKlf10) was 5′-GTCCTAAGGCTCAAGCCACC-3′. The sequence of siRNA against Pgc-1α (siPgc-1α) was generated as previously described [22].
Cell culture
Primary mouse hepatocytes were obtained from the livers of male C57BL/6J mice (8 weeks of age) and cultured as previously reported [23]. Mouse hepatocytes were infected with Ad-Klf10, Ad-gfp, Ad-shCtrl or Ad-shKlf10. for 36–48 h, and cells were then harvested for further analysis. For co-infection experiments, 24 h after infection with adenovirus expressing shPgc-1α (Ad-shPgc-1α), cells were co-infected with Ad-Klf10 or Ad-gfp for another 24 h. Cells were then harvested for further analysis.
RNA extraction and quantitative (q) real-time PCR
Total RNA was extracted with TRIzol (Invitrogen) from cells or pulverised liver tissues, and then reverse-transcribed to cDNA using a high-capacity cDNA reverse transcription kit (Applied Biosystems, Carlsbad, CA, USA). qPCR was performed as previously described [23]. mRNA levels for specific genes were normalised by β-actin mRNA levels. All primers are listed in ESM Table 2.
Western blot analysis
Proteins were extracted from pulverised livers or cultured hepatocytes in cell lysis buffer, 40–80 μg of protein was resolved using 10% SDS–polyacrylamide gel and separated proteins were eletrotransferred to polyvinylidene difluoride membranes. Western blot assays were performed using antibodies specific for rabbit anti-PGC-1α (Cell Signaling Technology, Boston, MA, USA), rabbit anti-KLF10 (Cell Signaling Technology), mouse anti-sterol regulatory element binding transcription factor (SREBP)-1 (Santa Cruz Biotechnology, Santa Cruz, CA, USA) and mouse anti-β-tubulin (Beijing ComWin Biotech Co, Beijing, China). See ESM Table 3 for antibody details.
Glucose output assay
Primary mouse hepatocytes were seeded in 6-well plates and then infected with the indicated adenovirus. At 36 h after infection, cells were washed three times with PBS and maintained in 2 ml/well of phenol-red-free, glucose-free DMEM containing 1 μmol/l dexamethasone, 2 mmol/l pyruvate, 20 mmol/l lactate and 10 μmol/l forskolin for 3–6 h. The glucose concentration in the medium was measured using an Amplex Red Glucose/Glucose Oxidase Assay Kit (Applygen Technologies, Beijing, China). Cells were lysed, and the protein concentration was determined (Bio-Rad, Hercules, CA, USA) for each lysate. The glucose output rate was normalised by the cellular protein content.
Analytical procedures and chemicals
Serum concentrations of glucose, triacylglycerol, total cholesterol, NEFA, alanine transaminase and aspartate aminotransferase were determined using an automated Monarch device (Instrumentation Laboratory, Lexington, MA, USA) at the clinical laboratory of Peking Union Medical College Hospital, Beijing, China. Hepatic concentrations of triacylglycerol and total cholesterol were measured using a colorimetric diagnostic kit (Applygen Technologies).
GTT and pyruvate tolerance test (PTT)
For in vivo infections, the indicated adenoviruses were delivered by tail-vein injection into C57BL/6J, db/db or DIO mice. Five days after injection, mice were i.p. injected with d-glucose (1–2 g/kg) or pyruvate sodium (0.5–1.5 g/kg) after 16 h fasting. Blood glucose levels were measured from the tail vein using a glucose monitor (OneTouch Ultra; LifeScan, Milpitas, CA, USA) at 0, 15, 30, 45, 60, 90 and 120 min.
Luciferase reporter gene assay
HepG2 cells (American Type Culture Collection, Manassas, VA, USA) were grown in 24-well plates using Dulbecco′s Modified Eagle Medium containing 10% (vol./vol.) FBS (Invitrogen). Luciferase reporter genes were then cotransfected into cells, together with the indicated expression plasmids. The Ramlila luciferase expression vector pCMV-RL-TK (Promega) was used as an internal control. After 48 h, cells were harvested and assessed for luciferase activity using the Dual Luciferase Reporter Assay System (Promega). Relative luciferase activity was corrected for Renilla luciferase activity of pCMV-RL-TK, and normalised to the activity of the control.
Chromatin immunoprecipitation (ChIP) assay
Pulverised liver tissues from C57BL/6J, db/m or db/db mice were lysed and sonicated as previously described [24]. The protein–DNA complexes were immunoprecipitated with mouse IgG antibody (control) or anti-KLF10 antibody. The promoter region of Pgc-1α was amplified by PCR or qPCR using the following primer pair: 5′-TGTTGCCTTCAAACACTCCTCT-3′ as a forward primer and 5′-ACACAGTAACTACACCCTTGCC-3′ as a reverse primer.
Statistical analysis
Data are presented as means ± SEM of more than three independent experiments. Statistical analysis was performed using Student’s t test (*p < 0.05, **p < 0.01, ***p < 0.001).
Results
Nutritional regulation of Klf10 gene expression in mouse livers
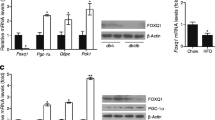
To identify novel transcriptional factors involved in dysfunctional hepatic glucose and lipid metabolism in obesity and diabetes, we performed mRNA microarray analysis on the livers of db/db mice. Interestingly, our preliminary mRNA microarray data indicated that hepatic Klf10 expression levels are increased in db/db mice compared with control wild-type C57BL/6J mice. We further detected hepatic expression of Klf10 by real-time PCR in different mouse models, including db/db, DIO and ob/ob mice. We found that hepatic Klf10 expression levels in these mice with metabolic disease were higher than those in their respective control mice (Fig. 1a–c).
Hepatic Klf10 gene expression is regulated by nutritional status in wild-type C57BL/6J mice and upregulated in diabetic, DIO and obese mice. (a–c) qPCR analysis and western blot analysis of hepatic Klf10 expression in (a) db/m control mice (white bar) and diabetic db/db mice (black bar), (b) C57BL/6J mice fed normal chow (control mice, white bar) or a high-fat diet for 16 weeks (HFD; DIO mice, black bar) and (c) C57BL/6J control mice (white bar) and obese ob/ob mice (black bar). (d) qPCR analysis of Klf10, Pgc-1α, Pck1 and G6pc mRNA expression in 8-week-old male C57BL/6J mice fed ad libitum (white bars) or fasted for 24 h (black bars). n = 4 mice per group. Data are means ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001
Next, we investigated whether hepatic Klf10 expression could be regulated by nutritional status in wild-type mice. We found that prolonged fasting (24 h) induced hepatic Klf10 gene expression in wild-type C57BL/6J mice (Fig. 1d). Meanwhile, we also observed that the expression levels of genes involved in gluconeogenesis, including Pgc-1α, Pck1 and G6pc, were markedly upregulated under prolonged fasting conditions (Fig. 1d). Taken together, these findings imply a strong correlation between Klf10 expression and gluconeogenesis in the liver.
KLF10 regulates a cellular gluconeogenic programme in primary hepatocytes
To explore the functional importance of KLF10 expression in the gluconeogenic programme in vitro, we generated recombinant Ad-Klf10. Overexpression of KLF10 in primary mouse hepatocytes induced by Ad-Klf10 treatment markedly increased the expression of Pgc-1α, Pck1 and G6pc (Fig. 2a). Correspondingly, the forced expression of KLF10 significantly stimulated glucose production in primary hepatocytes (Fig. 2b). In the fasted state, glucagons are secreted to stimulate hepatic gluconeogenesis. To test whether KLF10 is required for glucagon induction of gluconeogenic genes, we infected primary hepatocytes with Ad-shKlf10 expressing Klf10-specific shRNA and then treated the cells with forskolin, which mimics the effect of glucagon on increasing cellular cAMP levels. KLF10 knockdown was found to significantly decrease the expression of gluconeogenic genes and the stimulatory effects of forskolin on gluconeogenic genes, including Pgc-1α, Pck1 and G6pc (ESM Fig. 1a, Fig. 2c).
KLF10 regulates a cellular gluconeogenic programme in primary hepatocytes. (a) qPCR analysis of Pgc-1α, Pck1 and G6pc mRNA levels and western blot analysis of KLF10 and PGC-1α protein levels in mouse primary hepatocytes infected with Ad-gfp (white bars) or Ad-Klf10 (black bars). (b) Glucose production (glucose output) in primary hepatocytes treated as in (a). (c) qPCR analysis of mRNA levels of Pgc-1α, Pck1 and G6pc in mouse primary hepatocytes infected with Ad-shCtrl (control) or Ad-shKlf10 in the presence or absence of forskolin (white bars, Ad-shCtrl; black bars, Ad-shKlf10; light grey bars, Ad-shCtrl + forskolin; dark grey bars, Ad-shKlf10 + forskolin). (d) qPCR analysis of mRNA levels of Pck1 and G6pc in mouse primary hepatocytes infected with the indicated adenovirus (white bars, Ad-gfp + Ad-shCtrl; black bars, Ad-Klf10 + Ad-shCtrl; grey bars, Ad-Klf10 + adenovirus expressing shPgc-1α). Data are means ± SEM of three or four replicates. *p < 0.05, **p < 0.01, ***p < 0.001
Since both Pck1 and G6pc are downstream target genes of PGC-1α, we next explored whether the effects of KLF10 on the expression of Pck1 and G6pc are dependent on PGC-1α. We treated hepatocytes with Ad-Klf10 and Ad-shPgc-1α, which effectively expresses PGC-1α-specific shRNA. We found that knockdown of PGC-1α almost abolished the effects of KLF10 on the expression of gluconeogenic genes (ESM Fig. 1b, Fig. 2d).
In addition, we found that KLF10 overexpression in primary mouse hepatocytes also increased cellular triacylglycerol concentration and induced the expression of lipogenic genes, including Srebp-1c (also known as Srebf1), Fas and Acc1 (also known as Acaca) (ESM Fig. 2).
Hepatic overexpression of KLF10 in wild-type C57BL/6J mice increases blood glucose levels and impairs glucose tolerance
Based on the above results, it is reasonable to speculate that KLF10 might affect hepatic gluconeogenesis. To investigate the effect of KLF10 on glucose metabolism in vivo, we injected Ad-Klf10 into C57BL/6J mice via the tail vein. The injection of Ad-Klf10 significantly increased hepatic expression of KLF10 (Fig. 3a), but did not affect KLF10 expression in the muscle or white adipose tissue examined (data not shown). Consistent with the above results, hepatic overexpression of KLF10 increased the expression levels of gluconeogenic genes, including Pgc-1α, Pck1 and G6pc (Fig. 3a). Furthermore, fasting blood glucose levels in Ad-Klf10-infected mice were higher than those of Ad-gfp-infected control mice (Fig. 3b). We also performed GTTs to determine the effect of hepatic KLF10 overexpression on glucose tolerance. The GTT experiments indicated that overexpression of KLF10 in the liver impaired glucose tolerance (Fig. 3c). PTT experiments confirmed that the stimulation of gluconeogenesis in the liver by hepatic KLF10 overexpression contributed to the high glucose levels (Fig. 3d).
Overexpression of KLF10 in C57BL/6J mice increases blood glucose levels and impairs glucose tolerance. Male C57BL/6J mice were injected with Ad-gfp (white bars/black circles) or Ad-Klf10 (black bars/squares). Five to eight days after infection, the mice were killed for further analysis. (a) qPCR analysis of mRNA levels and western blot analysis of protein levels of hepatic KLF10 and gluconeogenic genes/proteins in Ad-gfp- or Ad-Klf10-injected C57BL/6J mice. (b) Blood glucose levels of 6 h-fasted C57BL/6J mice on day 8 after injection with the indicated adenovirus. (c, d) Blood glucose concentrations during (c) the GTT and (d) PTT in C57BL/6J mice infected with the indicated adenovirus. n = 4–6 mice per group. Data are means ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001
In addition, we also investigated the influence of hepatic KLF10 overexpression in C57BL/6J mice on lipogenesis, since KLF10 was found to induce the expression of lipogenic genes in primary hepatocytes (ESM Fig. 2). We found that Ad-Klf10-infected mice had increased expression of Srebp-1c, Fas and Acc1 (ESM Fig. 3). Correspondingly, KLF10 overexpression in the liver of C57BL/6J mice increased hepatic and serum triacylglycerol levels, but was not associated with significant changes in body weight, liver/body-weight ratio or serum cholesterol levels (ESM Table 4).
Taken together, these data suggest that upregulation of KLF10 expression in the liver of C57BL/6J mice induces hepatic gluconeogenesis, resulting in hyperglycaemia and glucose intolerance.
Hepatic silencing of KLF10 ameliorates hyperglycaemia and glucose intolerance in db/db diabetic and DIO mice
To further confirm the physiological roles of KLF10 in hepatic glucose metabolism, we injected Ad-shKlf10 into db/db mice via the tail vein. The injection of Ad-shKlf10 significantly reduced the hepatic expression of KLF10 compared with injection of Ad-shCtrl (Fig. 4a), without affecting KLF10 expression in skeletal muscle and abdominal white adipose tissue (data not shown). Consistent with observations in primary hepatocytes, knockdown of KLF10 markedly inhibited the hepatic expression of gluconeogenic genes (Fig. 4a). Ad-shKlf10-infected mice displayed significantly decreased fasting blood glucose levels compared with Ad-shCtrl-infected control mice (Fig. 4b). Subsequent GTT experiments showed that hepatic KLF10 knockdown improved glucose intolerance (Fig. 4c), while PTT experiments confirmed that the inhibition of gluconeogenesis by hepatic KLF10 knockdown resulted in decreased blood glucose levels in db/db mice (Fig. 4d). Moreover, we also found that hepatic knockdown of KLF10 in db/db mice inhibited the expression of lipogenic genes (ESM Fig. 4) and reduced hepatic triacylglycerol levels, although serum triacylglycerol levels, liver weight/body-weight ratio, and serum and hepatic cholesterol levels remained similar to those in control mice (ESM Table 5).
Knockdown of KLF10 in db/db mice decreases blood glucose levels and improves glucose tolerance. Male db/db mice were injected with Ad-shCtrl (white bars/black circles) or Ad-shKlf10 (black bars/squares). Five to eight days after infection, the mice were killed for further analysis. (a) qPCR analysis of mRNA levels and western blot analysis of protein levels of KLF10 and gluconeogenic genes/proteins in the livers of db/db mice injected with Ad-shCtrl or Ad-shKlf10. (b) Blood glucose levels of 6 h-fasted db/db mice on day 8 after injection with the indicated adenovirus. (c, d) Blood glucose concentrations during (c) the GTT and (d) PTT in db/db mice infected with the indicated adenovirus. n = 4–5 mice per group. Data are means ± SEM. *p < 0.05, **p < 0.01
Similar results were observed in DIO mice (Fig. 5, ESM Fig. 5, ESM Table 6). Hepatic knockdown of KLF10 in DIO mice suppressed the expression of gluconeogenic genes, decreased blood glucose levels and improved glucose tolerance (Fig. 5). KLF10 knockdown also inhibited the expression of lipogenic genes (ESM Fig. 5), leading to reduced hepatic and serum triacylglycerol levels (ESM Table 6). These data indicated that KLF10 exerts a critical regulatory effect on glucose and lipid metabolism.
Knockdown of KLF10 in DIO mice decreases blood glucose levels and improves glucose tolerance. Male DIO mice were injected with Ad-shCtrl (white bars/circles) or Ad-shKlf10 (black bars/squares). Five to eight days after infection, the mice were killed for further analysis. (a) qPCR analysis of mRNA levels and western blot analysis of protein levels of hepatic KLF10 and gluconeogenic genes/proteins in Ad-shCtrl- or Ad-shKlf10-injected DIO mice. (b) Blood glucose levels of 6 h-fasted DIO mice on day 8 after injection with the indicated adenovirus. (c, d) Blood glucose concentrations during (c) the GTT and (d) PTT in DIO mice infected with the indicated adenovirus. n = 4–5 mice per group. Data are means ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001
KLF10 activates transcription of Pgc-1α, a gluconeogenic gene, via binding to its promoter
The above data suggest that KLF10 is an important regulator of gluconeogenic genes, including Pgc-1α and its downstream target genes Pck1 and G6pc. Since KLF10 is a transcription factor, we wondered whether KLF10 protein binds to the Pgc-1α gene promoter. The promoter region of the Pgc-1α gene (2525 bp) was cloned and fused to a luciferase reporter gene (−2525Luc). In addition, we generated a series of luciferase reporter constructs containing shorter fragments of the Pgc-1α gene promoter (−729Luc, −171Luc and −34Luc). The luciferase reporter gene assay showed that overexpression of KLF10 activated transcription of −2525Luc, −729Luc and −171Luc in HepG2 cells (Fig. 6a). However, the stimulatory effects of KLF10 were abolished when the promoter region was further truncated to −34 bp (−34Luc) (Fig. 6a), suggesting the sequence between −171 and −34 bp mediates the effects of KLF10 on Pgc-1α gene transcription.
KLF10 stimulates transcription of the Pgc-1α gene by directly binding to its promoter. (a) Luciferase reporter gene assay in HepG2 cells transfected with the indicated plasmids. A series of truncated Pgc-1α promoters fused to the luciferase reporter gene were cotransfected into HepG2 cells, together with pcDNA3.1 (control, white bars) or KLF10-expression plasmids (black bars). Luc, luciferase; RLA, relative luciferase activity. (b) ChIP analysis on liver tissues of C57BL/6J mice to assess endogenous KLF10 occupancy of the Pgc-1α promoter. DL2000, DNA marker purchased from Takara Bio (Shiga, Japan). (c) ChIP analysis on liver tissues of db/m (white bars) and db/db (black bars) mice, and qPCR assessed for endogenous KLF10 occupancy of the Pgc-1α promoter. n = 3 mice per group. Results are means ± SEM of three or four replicates. *p < 0.05, **p < 0.01, ***p < 0.001. IP, immunoprecipitation
Next, we performed ChIP assays to investigate whether endogenous KLF10 protein could directly bind to the Pgc-1α promoter in vivo. We found that the Pgc-1α promoter fragment containing a KLF10 binding site (from −21 bp to −182 bp) could be amplified from the precipitates obtained when using anti-KLF10 antibody, but not when using normal rabbit IgG (negative control) in the liver tissue lysate of C57BL/6J mice (Fig. 6b). In addition, ChIP–qPCR analyses confirmed the presence of a greater amount of endogenous KLF10 protein associated with the Pgc-1α promoter in the liver of db/db mice compared with db/m mice (Fig. 6c). These observations suggested that KLF10 increases the transcription of Pgc-1α via direct binding to its promoter region.
Discussion
KLFs have been shown to be involved in the regulation of critical cellular processes, including cell proliferation, development, apoptosis and differentiation [14, 15]. In the past few years, accumulating evidence has suggested that multiple KLFs are involved in hepatic or systemic glucose metabolism [15]. For example, overexpression of KLF7 has been reported to inhibit the expression and glucose-induced secretion of insulin in a pancreatic beta-cell line [25]. KLF15 regulates hepatic gluconeogenesis and is an important target of metformin [26, 27]. In addition, we have previously demonstrated that hepatic KLF11 expression is regulated by nutritional states, and that KLF11 is an important regulator of hepatic lipid and glucose metabolism [23, 28].
KLF10 is highly expressed in the liver, and its expression is regulated by nutritional signals (high glucose levels) and the transcription factor carbohydrate-responsive element-binding protein (ChREBP). Consistent with our data, a previous report also showed that hepatic expression of KLF10 is higher in ob/ob mice compared with C57BL/6J mice [18, 29]. In muscle, KLF10 expression is regulated by insulin [30]. Moreover, Klf10 gene variants have been associated with susceptibility to type 2 diabetes [17, 18]. These studies suggest that KLF10 might regulate systemic metabolism.
In the current study, hepatic KLF10 expression was regulated by nutritional status in wild-type C57BL/6J mice. The secretion of glucagon and glucocorticoids increases to activate hepatic gluconeogenesis under fasting conditions, while insulin levels drop [1, 31]. Thus, we speculated that glucagon and glucocorticoids might induce KLF10 in primary hepatocytes. We treated primary hepatocytes with forskolin and dexamethasone, which mimic the effects of glucagon and glucocorticoids, respectively. Although the treatment of cells with these compounds markedly induced the expression of gluconeogenic genes such as Pck1 and G6pc, they did not influence the expression of Klf10 (data not shown). Additional studies are required to identify potential triggers of KLF10 activation under fasting and pathophysiological conditions.
Guillaumond et al reported that male mice with global knockout of Klf10 displayed postprandial and fasting hyperglycaemia, whereas Klf10-deficient female mice displayed higher plasma triacylglycerol concentrations [29]. Thus, the effects of KLF10 on metabolism are influenced by sex. Possibly these different metabolic phenotypes might be attributed to sex-specific hormones altered by KLF10 deficiency in other tissues, since KLF10 is ubiquitously expressed in multiple tissues, including the kidneys, heart, bone, muscle and fat [29]. However, the exact molecular mechanism underlying this discrepancy remains unclear.
In the present study, we explored the role of KLF10 in regulating hepatic glucose metabolism with Ad-Klf10 or Ad-shKlf10, which led to an acute change in hepatic KLF10 expression. We found that liver-specific overexpression of KLF10 increased Pgc-1α transcription, which resulted in hyperglycaemia and impaired glucose tolerance in C57BL/6J mice. Knockdown of KLF10 in the liver attenuated hyperglycaemia and glucose intolerance in db/db or DIO mice. In addition, our ChIP data confirm that endogenous KLF10 protein can directly bind to the Pgc-1α promoter. Our data suggest that Klf10 is a gluconeogenic gene. Notably, the metabolic phenotypes observed in the present study resulted from short-term alterations in KLF10 expression in liver, while hyperglycaemia in global Klf10-deficient male mice resulted from long-term effects of KLF10 deficiency, which may be attributed to its roles in other tissues.
In addition to glucose metabolism, we also studied the role of KLF10 in regulating lipid metabolism in hepatocytes. We found that overexpression of KLF10 in mouse primary hepatocytes increased lipogenesis. Consistently, overexpression of KLF10 in the livers of normal (wild-type) mice elevated hepatic and serum triacylglycerol content. However, hepatic knockdown of KLF10 decreased hepatic and serum triacylglycerol in DIO mice. Furthermore, our luciferase reporter gene assay suggests that KLF10 does not activate transcription of the Srebp-1c gene (data not shown), indicating that KLF10 affects the expression of lipogenic genes via other mechanisms, such as affecting mRNA stability or other factors influencing Srebp-1c. Further studies are required to clarify this.
KLF15 has been reported to control gluconeogenesis by regulating amino-acid-degrading enzymes to provide gluconeogenic substrate [26]. Considering that KLF10 and KLF15 have similar structures and gluconeogenic functions, we also examined the expression levels of alanine transaminase 1 (ALT1), 4-hydroxyphenylpyruvic acid dioxygenase (HPD) and proline dehydrogenase (ProDH), which catabolise alanine, tyrosine and proline, respectively. However, KLF10 overexpression did not markedly stimulate the genes encoding these proteins (data not shown), which differs from findings for KLF15 [27]. In addition, KLF15 does not affect Pck1 expression [27], which is different from KLF10 as shown in the present study. Thus, it appears that these two transcription factors regulate hepatic gluconeogenesis through different mechanisms.
In summary, our study suggests that increased KLF10 expression might be an underlying factor for increased hepatic gluconeogenesis in diabetic mice, contributing to the development of diabetes. Thus, modulation of KLF10 may be a novel therapeutic approach for the treatment of diabetes.
Abbreviations
- Ad-gfp :
-
Adenovirus expressing green fluorescent protein
- Ad-Klf10 :
-
Adenovirus expressing Krüppel-like factor 10
- Ad-shCtrl:
-
Control adenovirus expressing short-hairpin RNA against luciferase
- Ad-shKlf10 :
-
Adenovirus expressing short-hairpin RNA against Krüppel-like factor 10
- Ad-shPgc-1α :
-
Adenovirus expressing short-hairpin RNA against peroxisome proliferator-activated receptor, gamma, coactivator 1, alpha
- ChIP:
-
Chromatin immunoprecipitation
- CREB:
-
Cyclic AMP response element-binding protein
- DIO:
-
Diet-induced obese
- KLF:
-
Krüppel-like factor
- PGC-1α:
-
Peroxisome proliferator-activated receptor, gamma, coactivator 1α
- PTT:
-
Pyruvate tolerance test
- qPCR:
-
Quantitative PCR
- shRNA:
-
Short-hairpin RNA
- siRNA:
-
Small interfering RNA
- SREBP:
-
Sterol regulatory element-binding transcription factor
References
Yoon JC, Puigserver P, Chen G et al (2001) Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature 413:131–138
Pilkis SJ, Granner DK (1992) Molecular physiology of the regulation of hepatic gluconeogenesis and glycolysis. Annu Rev Physiol 54:885–909
Zhou Y, Jiang L, Rui L (2009) Identification of MUP1 as a regulator for glucose and lipid metabolism in mice. J Biol Chem 284:11152–11159
Saltiel AR, Kahn CR (2001) Insulin signalling and the regulation of glucose and lipid metabolism. Nature 414:799–806
Herzig S, Long F, Jhala US et al (2001) CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature 413:179–183
Puigserver P, Spiegelman BM (2003) Peroxisome proliferator-activated receptor-γ coactivator 1α (PGC-1α): transcriptional coactivator and metabolic regulator. Endocr Rev 24:78–90
Koo S-H, Flechner L, Qi L et al (2005) The CREB coactivator TORC2 is a key regulator of fasting glucose metabolism. Nature 437:1109–1111
Puigserver P, Rhee J, Donovan J et al (2003) Insulin-regulated hepatic gluconeogenesis through FOXO1–PGC-1α interaction. Nature 423:550–555
Lin J, Handschin C, Spiegelman BM (2005) Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab 1:361–370
Li X, Monks B, Ge Q, Birnbaum MJ (2007) Akt/PKB regulates hepatic metabolism by directly inhibiting PGC-1α transcription coactivator. Nature 447:1012–1016
Oberkofler H, Schraml E, Krempler F, Patsch W (2003) Potentiation of liver X receptor transcriptional activity by peroxisome-proliferator-activated receptor gamma co-activator 1 alpha. Biochem J 371:89–96
Rhee J, Inoue Y, Yoon JC et al (2003) Regulation of hepatic fasting response by PPARγ coactivator-1α (PGC-1): requirement for hepatocyte nuclear factor 4α in gluconeogenesis. Proc Natl Acad Sci U S A 100:4012–4017
Nakae J, Biggs WH 3rd, Kitamura T et al (2002) Regulation of insulin action and pancreatic β-cell function by mutated alleles of the gene encoding forkhead transcription factor Foxo1. Nat Genet 32:245–253
Kaczynski J, Cook T, Urrutia R (2003) Sp1- and Kruppel-like transcription factors. Genome Biol 4:206
McConnell BB, Yang VW (2010) Mammalian Kruppel-like factors in health and diseases. Physiol Rev 90:1337–1381
Subramaniam M, Hawse JR, Rajamannan NM, Ingle JN, Spelsberg TC (2010) Functional role of KLF10 in multiple disease processes. Biofactors 36:8–18
Gutierrez-Aguilar R, Benmezroua Y, Balkau B et al (2007) Minor contribution of SMAD7 and KLF10 variants to genetic susceptibility of type 2 diabetes. Diabete Metab 33:372–378
Iizuka K, Takeda J, Horikawa Y (2011) Kruppel-like factor-10 is directly regulated by carbohydrate response element-binding protein in rat primary hepatocytes. Biochem Biophys Res Commun 412:638–643
Wahab NA, Weston BS, Mason RM (2005) Modulation of the TGFβ/Smad signaling pathway in mesangial cells by CTGF/CCN2. Exp Cell Res 307:305–314
Kim JK, Lee KS, Chang HY, Lee WK, Lee JI (2014) Progression of diet induced nonalcoholic steatohepatitis is accompanied by increased expression of Kruppel-like-factor 10 in mice. J Transl Med 12:186
Luo J, Deng ZL, Luo X et al (2007) A protocol for rapid generation of recombinant adenoviruses using the AdEasy system. Nat Protoc 2:1236–1247
Koo SH, Satoh H, Herzig S et al (2004) PGC-1 promotes insulin resistance in liver through PPAR-α-dependent induction of TRB-3. Nat Med 10:530–534
Zhang H, Chen Q, Yang M et al (2013) Mouse KLF11 regulates hepatic lipid metabolism. J Hepatol 58:763–770
Wang R, Kong X, Cui A et al (2010) Sterol-regulatory-element-binding protein 1c mediates the effect of insulin on the expression of Cidea in mouse hepatocytes. Biochem J 430:245–254
Kawamura Y, Tanaka Y, Kawamori R, Maeda S (2006) Overexpression of Kruppel-like factor 7 regulates adipocytokine gene expressions in human adipocytes and inhibits glucose-induced insulin secretion in pancreatic β-cell line. Mol Endocrinol 20:844–856
Takashima M, Ogawa W, Hayashi K et al (2010) Role of KLF15 in regulation of hepatic gluconeogenesis and metformin action. Diabetes 59:1608–1615
Gray S, Wang B, Orihuela Y et al (2007) Regulation of gluconeogenesis by Kruppel-like factor 15. Cell Metab 5:305–312
Zhang H, Chen Q, Jiao T et al (2014) Involvement of KLF11 in hepatic glucose metabolism in mice via suppressing of PEPCK-C expression. PLoS One 9:e89552
Guillaumond F, Grechez-Cassiau A, Subramaniam M et al (2010) Kruppel-like factor KLF10 is a link between the circadian clock and metabolism in liver. Mol Cell Biol 30:3059–3070
Coletta DK, Balas B, Chavez AO et al (2008) Effect of acute physiological hyperinsulinemia on gene expression in human skeletal muscle in vivo. Am J Physiol Endocrinol Metab 294:E910–E917
Quinn PG, Yeagley D (2005) Insulin regulation of PEPCK gene expression: a model for rapid and reversible modulation. Curr Drug Targets Immune Endocr Metabol Disord 5:423–437
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Funding
This work was supported by the National Natural Science Foundation of China (grant nos. 81471049, 81670749, 81700768 and 81730024).
Duality of interest
The authors declare that there is no duality of interest associated with this manuscript.
Contribution statement
YC contributed to the conception and design of the study. XY and QC contributed to the study design and acquisition and analysis of data. LS, HZ, LY, XC, YG and FF contributed to the analysis and interpretation of data. XY, QC and YC contributed to drafting or revising the article. All authors critically revised the manuscript and approved the final version. YC is the guarantor of this work.
Electronic supplementary material
ESM
(PDF 697 kb)
Rights and permissions
About this article
Cite this article
Yang, X., Chen, Q., Sun, L. et al. KLF10 transcription factor regulates hepatic glucose metabolism in mice. Diabetologia 60, 2443–2452 (2017). https://doi.org/10.1007/s00125-017-4412-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-017-4412-2