Abstract
Aims/hypothesis
Type 2 diabetes is a chronic metabolic disorder associated with devastating microvascular complications. Genome-wide association studies have identified more than 60 genetic variants associated with type 2 diabetes and/or glucose and insulin traits, but their role in the progression of diabetes is not established. The aim of this study was to explore whether these variants were also associated with the development of nephropathy in patients with type 2 diabetes.
Methods
We studied 28 genetic variants in 2,229 patients with type 2 diabetes from the local Malmö Scania Diabetes Registry (SDR) published during 2007–2010. Diabetic nephropathy (DN) was defined as micro- or macroalbuminuria and/or end-stage renal disease. Estimated glomerular filtration rate (eGFR) was assessed using the MDRD-4 formula. Replication genotyping of rs1531343 was performed in diabetic (Steno type 2 diabetes [n = 345], Genetics of Diabetes Audit and Research in Tayside Scotland [Go-DARTS] [n = 784]) and non-diabetic (Malmö Preventive Project [n = 2,523], Botnia study [n = 2,247]) cohorts.
Results
In the SDR, HMGA2 single-nucleotide polymorphism rs1531343 was associated with DN (OR 1.50, 95% CI 1.20, 1.87, p = 0.00035). In the combined analysis totalling 3,358 patients with type 2 diabetes (n = 1,233 cases, n = 2,125 controls), carriers of the C-allele had a 1.45-fold increased risk of developing nephropathy (95% CI 1.20, 1.75, p = 0.00010). Furthermore, the risk C-allele was associated with lower eGFR in patients with type 2 diabetes (n = 2,499, β ± SEM, −3.7 ± 1.2 ml/min, p = 0.002) and also in non-diabetic individuals (n = 17,602, β ± SEM, −0.008 ± 0.003 ml/min (log e ), p = 0.006).
Conclusions/interpretation
These data demonstrate that the HMGA2 variant seems to be associated with increased risk of developing nephropathy in patients with type 2 diabetes and lower eGFR in both diabetic and non-diabetic individuals and could thus be a common denominator in the pathogenesis of type 2 diabetes and kidney complications.
Similar content being viewed by others
Introduction
Type 2 diabetes is a metabolic disorder characterised by impaired insulin secretion and action that ultimately lead to chronic hyperglycaemia. Chronic hyperglycaemia is, in turn, associated with increased risk of progression to microvascular complications (nephropathy and retinopathy) of diabetes [1]. About 35% of patients with type 2 diabetes develop diabetic nephropathy (DN) [2]. Diabetes is also the most common cause of end-stage renal disease (ESRD) and the need for dialysis or kidney transplantation, which are associated with increased mortality [3, 4]. Several studies have demonstrated the preventive effect of controlling blood glucose on developing microvascular complications of diabetes [5–7].
DN is characterised by structural damage to the kidney, which results in leakage of albumin in urine. Both increased micro- (AER 20–200 μg/min or albumin/creatinine ratio >2.5 and >3.5 mg/mmol in men and women, respectively) and macroalbuminuria (AER >200 μg/min) are strongly associated with risk of morbidity and mortality from cardiovascular diseases [8, 9]. The GFR describes the flow rate of filtered fluid by the kidney and is used for assessment of renal function. Since direct measurements of GFR are tedious, the estimated GFR (eGFR) is often predicted from serum creatinine (S-creatinine), age, sex and ethnicity [10]. Clustering of DN within ethnic groups and families [11] indicates existing genetic predisposition to renal pathology [11]. Genome-wide associated studies (GWASs) have been shown to be an unbiased approach to identify genetic susceptibility loci for a number of diseases. Currently there are more than 60 common genetic variants that have been associated with type 2 diabetes and/or glucose or insulin levels using GWASs [12]. However, whether these variants also associated with the development of complications of diabetes is not established. In this study we explored whether variants influencing type 2 diabetes and/or glycaemic traits were also associated with the development of nephropathy in patients with type 2 diabetes.
Methods
Participants
Scania Diabetes Registry
The Scania Diabetes Registry (SDR) was initiated in Malmö (south of Sweden) in 1996; the majority of patients regularly attended the Department of Endocrinology, Skåne University Hospital. The aim of the study was to find factors associationed with the development of complications using biomarkers and genetic markers [13]. Microalbuminuria was defined as at least two out of three consecutive measurements with AER ≥ 20 < 200 μg/min. Macroalbuminuria was defined as at least one measurement with AER ≥200 μg/min or ≥300 mg/24 h. ESRD was defined as eGFR ≤15 ml/min or dialysis or kidney transplantation. S-creatinine was determined using an enzymatic colorimetric method (Cobas NPU04998; Roche Diagnostic, Basel, Switzerland). Urine albumin was determined using immunonephelometry (Beckman Instruments, Brea, CA, USA) until 1998 and thereafter using an immunoturbimetric method (Beckman Coulter; Beckman Instruments, Brea, CA, USA) [14]. eGFR was estimated using the MDRD-4 formula. Nephropathy was defined as micro- or macroalbuminuria and/or ESRD.
Steno type 2 diabetes cohort
In the Steno cohort patients with type 2 diabetes (n = 345) were included as either cases with DN or controls without nephropathy collected as part of a European case–control study collaboration [15, 16]. The study design included nephropathy with inclusion criteria of albuminuria >300 mg/l and presence of diabetic retinopathy to ensure that albuminuria was the consequence of DN rather than a non-diabetic glomerulopathy [17]. Urinary albumin was determined using an enzyme immunoassay. S-creatinine was determined using a modified Jaffe’s method.
Genetics of Diabetes Audit and Research in Tayside Scotland
The Genetics of Diabetes Audit and Research in Tayside Scotland (Go-DARTS) database includes prescription and biochemistry information and clinical phenotypes of all patients with diabetes within Tayside, Scotland, from 1992 onwards. The Go-DARTS study is a joint initiative of the Department of Medicine and the Medicines Monitoring Unit (MEMO) at the University of Dundee. A total of 3,800 patients with type 2 diabetes from the Go-DARTS cohort were genotyped on the Affymetrix 6.0 single nucleotide polymorphism (SNP) array (Affymetrix, High Wycombe, UK) and linked with the Go-DARTS database. DN was defined as macroalbuminuria (albumin–creatinine ratio [ACR] ≥25 mg/mmol for men and ACR ≥35 for women) [18].
In all diabetic cohorts (SDR, Steno and Go-DARTS) controls were defined having normoalbuminuria.
Malmö Preventive Project
The Malmö Preventive Project (MPP) is a population-based study from the city of Malmö in southern Sweden. The study started with 33,346 participants in total, of which 25,677 eligible persons participated in a health screening visit during 1974–1992 aiming at investigating large strata of the adult population to find high-risk individuals for preventive interventions. Blood samples were collected for measurements of fasting blood glucose and lipid concentrations. S-creatinine was determined with Jaffe’s alkaline picrate method [19]. During 2002–2006, 17,284 persons participated in the re-screening visit and blood samples were taken for genetic analyses [20]. In our study we included 15,066 non-diabetic individuals with available baseline S-creatinine data, of them 2,536 with available S-creatinine data at follow-up.
Botnia Study
The Botnia study is a family-based study from the western coast of Finland that started in 1990, aiming to identify genes that increase susceptibility to type 2 diabetes [21]. In the present study, we included 2,247 non-diabetic participants from the Botnia study with available eGFR measurements.
For all studies the protocols were approved by local ethics committees, and informed consent was obtained from all participants.
Assessment of renal function
Morning ACR and/or overnight AER were used for classification of nephropathy in patients with type 2 diabetes. In addition, as a marker of renal function, we used eGFR calculated using the MDRD-4 formula: eGFR = 186 × S-creatinine−1.54 × Age−0.203 × (1.210 if black) × (0.742 if female sex) [16, 22].
Genotyping
We genotyped 28 known SNPs associated with type 2 diabetes/glycaemic traits published during 2006–2010 [23–27] in 2,229 patients with type 2 diabetes from SDR using the Mass Extend Mass ARRAY system (Sequenom, San Diego, CA, USA). Out of 28 SNPs 26 were in Hardy–Weinberg equilibrium with the call rate ranging from 90% to 100% (electronic supplementary material [ESM] Table 1). Replication of the HMGA2 variants SNP rs1531343 in the Steno, MPP and Botnia cohorts was performed using the TaqMan allelic discrimination assay with an ABI 7900HT sequence detection system (Applied Biosystems, Foster City, CA, USA). In Go-DARTS the SNP rs1531343 was directly genotyped on the Affymetrix 6.0 SNP array.
Statistical analysis
The OR for developing nephropathy was analysed using logistic regression, adjusted for sex and diabetes duration as well as with and without HbA1c. Univariate linear regression was used to study the association of SNP rs1531343 (genotypic additive model) with eGFR adjusted for sex, diabetes duration and HbA1c in patients with type 2 diabetes (SDR and Steno) and with log e transformed eGFR (log e eGFR) adjusted for age, sex and BMI in non-diabetic individuals (MPP and Botnia). HbA1c in SDR was calculated as mean value of HbA1c during a mean follow-up period of 10.9 years. Diabetes duration was calculated from the age at onset of diabetes until development of nephropathy for cases and from age at onset of diabetes until last visit for controls. Bonferroni correction for multiple comparisons was used with an adjusted p level = 0.0018 (0.05/28). Statistical analyses were carried out using Statistical Package for the Social Sciences version 17.0 (SPSS, Chicago, IL, USA). Fixed-effects meta-analysis was performed by Meta-Analysis Package for R (Metafor 1.6-0; http://www.metafor-project.org/). Power calculations were performed using Quanto (Quanto Version 1.2.4; http://hydra.usc.edu/gxe).
Results
Clinical characteristics of the study participants are shown in Table 1. In the SDR, 947 (42%) patients progressed to nephropathy during a 10.2-year period. In the Steno study, there were 172 (50%) nephropathy cases with mean diabetes duration of 15.5 years. In the Go-DARTS cohort, 114 (15%) patients progressed to nephropathy during a 10.8-year period.
The prevalence and/or incidence of DN in men were significantly higher than in women (Table 1). In all studies, diabetic patients with nephropathy had higher HbA1c (SDR, 7.1% [65.6 mmol/mol] vs 6.6% [60.47 mmol/mol], p = 4.9 × 10−24; Steno, 9.1% [76.6 mmol/mol] vs 8.6% [71.5 mmol/mol], p = 0.009; Go-DARTS, 8.0% [62.53 mmol/mol] vs 7.5% [56.07 mmol/mol], p = 0.0002) and in two of the studies patients with nephropathy were younger at onset of diabetes (SDR, 53.6 ± 12 vs 54.6 ± 12.1 years, p = 0.021; Steno, 45.7 ± 12.4 vs 47.5 ± 9.3 years, p = 0.124) compared with those without nephropathy.
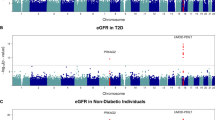
In the SDR, out of the 28 genetic loci analysed, the HMGA2 SNP rs1531343 was associated with increased risk of developing nephropathy after correction for multiple testing (ESM Table 2). The frequency of the minor C-allele of HMGA2 rs1531343 was significantly higher in patients with type 2 diabetes who had nephropathy (10.2% vs 6.9%, p = 9.8 × 10−5) as compared with those who did not. This was translated into a 1.5-fold increased risk of developing nephropathy (95% CI, 1.20, 1.87, p = 0.00035) adjusted for sex, diabetes duration and HbA1c. To replicate this finding, we validated this association in the additional available diabetic Steno and Go-DARTS cohorts. Although these cohorts had less power, similarly to SDR we observed that the frequency of the minor C-allele of HMGA2 rs1531343 was significantly higher in Steno (11.3% vs 6.6%, p = 0.031) and also tended to be higher in Go-DARTS (12.7% vs 10.9%, p = 0.41) patients with type 2 diabetes who had nephropathy as compared with those who did not. The combined analysis of SDR, Steno and Go-DARTS cohorts showed that carriers of the C-allele had a 1.45-fold increased risk of DN (95% CI, 1.20, 1.75, p = 0.00010) (Table 2). Additionally, we also explored whether HMGA2 rs1531343 would influence eGFR in patients with type 2 diabetes. We found that the C-allele of rs1531343 was also associated with lower eGFR (β ± SEM: −3.7 ± 1.2 ml/min, p = 0.002) (Table 3).
Next, we analysed 15,066 non-diabetic participants from the MPP study with available information on eGFR (Table 3). Notably, in the MPP the C-allele was associated with lower eGFR at baseline (β ± SEM: −0.007 ± 0.003 ml/min, p = 0.038) and remained lower after a 6.4-year follow-up period (β ± SEM: −0.021 ± 0.008 ml/min, p = 0.007). Similarly, in the non-diabetic participants of the Botnia study the risk C-allele was associated with lower eGFR (β ± SEM: −0.024 ± 0.012 ml/min, p = 0.040). Combined analysis of the MPP and Botnia studies strengthened this association (β ± SEM: −0.008 ± 0.003 ml/min, p = 0.006) (Table 3).
Discussion
In this study, we demonstrated that the risk C-allele of the common variant (rs1531343) in the HMGA2 loci predisposing to type 2 diabetes was also associated with progression to nephropathy in patients with type 2 diabetes and decline in renal function measured with eGFR in non-diabetic individuals.
Our observations are in line with a recently reported GWAS for kidney function (eGFR) [28]. The study included 67,093 participants of European ancestry from 20 predominantly population-based studies to identify new susceptibility loci for reduced renal function. The risk C-allele of SNP HMGA2 rs1531343 was also negatively associated with eGFR (p = 0.039) in this study, supporting our findings (https://intramural.nhlbi.nih.gov/labs/CF/Pages/CKDGenConsortium.aspx, accessed 08/08/2012) [28]. Notably, in a GWAS for DN in African-Americans, another variant (rs2358944), located 57 kb downstream of the HMGA2 rs1531343, was one of the strongest signals associated with diabetic ESRD, but also to a lesser extent with non-diabetic ESRD [29].
The high mobility group (HMG) protein family includes HMGA, HMGB and HMGN [30]. HMGA proteins are transcribed from two genes, HMGA1 and HMGA2 [31]. HMGA proteins are found in high levels in human embryonic stem cells and derived embryoid bodies in vitro [32]. HMGA2 is ascribed an important role in the control of stem-cell development and proliferation, and also high levels of HMGA2 is found in many benign, as well as malignant, tumours [33, 34]. HMG proteins are the most abundant non-histone DNA-binding chromatin factors in the eukaryotic nucleus involved in up- and downregulation of several genes [33].
Although the role of HMGA2 in diabetes or DN is still not fully understood, it was demonstrated that overexpression of HMGA2 was associated with formation of micropolycystic kidney suggesting involvement of HMGA2 in kidney development [35].
Diabetic kidney abnormalities are characterised mainly by hypertrophy, thickening of the glomerular basement membrane, tubular atrophy and interstitial fibrosis, which have been shown to lead to increased loss of renal function [33]. Recently, epithelial–mesenchymal transition (EMT) has been suggested as a novel pathway playing a key role in the pathogenesis of tubulointerstitial fibrosis in DN and blockage of EMT has been shown to delay progression to DN [33, 34]. Of note, transcriptomic analyses of TGF-β-stimulated EMT in NAMRU mouse mammary gland (NMuMG) cells identified HMGA2 as a main target involved in the control of epithelial differentiation [36]. TGF-β is a pro-sclerotic cytokine strongly associated with the development of fibrosis in DN [34]. The induction of cytokines, chemokines and growth factors, particularly TGFβ1 and connective tissue growth factor (CTGF), leads to glomerular basement membrane thickening, podocyte injury and mesangial matrix expansion. Thus changes in HMGA2 expression may ultimately increase the risk of development of irreversible glomerular sclerosis and tubulointerstitial fibrosis involved in the pathogenesis of DN. However, as for many genetic association studies, the observed phenotypic effect could be caused by one or several genetic variants in LD with the genotyped variant rather than the genotyped SNP itself. Thus, we cannot rule out that the effect is mediated by cis- or trans-regulatory genetic elements other than the HMGA2 gene.
Although in this study we had 96% power to detect an association of HMGA2 rs1531343 with nephropathy, the available replication cohorts were less powered. Also, the original discovery cohort still had limited power to detect an effect of other genetic loci with smaller effect sizes. Thus, we cannot exclude weaker associations and false-negative results. Larger studies will be needed to detect or exclude a role of these SNPs in DN. Additionally, given that long-term glycaemia and duration of diabetes are major determinants of progression to nephropathy, glycaemic control and longer duration of diabetes should be clearly applied, if available, to define patients without nephropathy in the design of future studies.
In conclusion, we demonstrate that an HMGA2 variant seems to be associated with increased risk of developing nephropathy in patients with type 2 diabetes and with lower eGFR in both diabetic and non-diabetic individuals and thus could represent a common denominator in the pathogenesis of type 2 diabetes and kidney complications. However, the biology of most genetic determinants influencing type 2 diabetes/glycaemic traits is most likely different from those involved in progression to complications of diabetes.
Abbreviations
- ACR:
-
Albumin–creatinine ratio
- DN:
-
Diabetic nephropathy
- EMT:
-
Epithelial–mesenchymal transition
- ESRD:
-
End-stage renal disease
- eGFR:
-
Estimated GFR
- Go-DARTS:
-
Genetics of Diabetes Audit and Research in Tayside Scotland
- GWAS:
-
Genome-wide association study
- HMG:
-
High mobility group
- MPP:
-
Malmö Preventive Project
- S-creatinine:
-
Serum creatinine
- SDR:
-
Scania Diabetes Registry
- SNP:
-
Single-nucleotide polymorphism
References
Chin MT, Pellacani A, Wang H et al (1998) Enhancement of serum-response factor-dependent transcription and DNA binding by the architectural transcription factor HMG-I(Y). J Biol Chem 273:9755–9760
Forbes JM, Fukami K, Cooper ME (2007) Diabetic nephropathy: where hemodynamics meets metabolism. Exp Clin Endocrinol Diabetes 115:69–84
Parving HH (2001) Diabetic nephropathy: prevention and treatment. Kidney Int 60:2041–2055
Harris MI, Flegal KM, Cowie CC et al (1998) Prevalence of diabetes, impaired fasting glucose, and impaired glucose tolerance in U.S. adults. The Third National Health and Nutrition Examination Survey, 1988–1994. Diabetes Care 21:518–524
Di Landro D, Catalano C, Lambertini D et al (1998) The effect of metabolic control on development and progression of diabetic nephropathy. Nephrol Dial Transplant 13(Suppl 8):35–43
The Diabetes Control and Complications Trial Research Group (1993) The effect of intensive treatment of diabetes on the development and progression of long-term complications in insulin-dependent diabetes mellitus. N Engl J Med 329: 977–986
UK Prospective Diabetes Study (UKPDS) Group (1998) Intensive blood-glucose control with sulphonylureas or insulin compared with conventional treatment and risk of complications in patients with type 2 diabetes (UKPDS 33). Lancet 352: 837–853
Valmadrid CT, Klein R, Moss SE, Klein BE (2000) The risk of cardiovascular disease mortality associated with microalbuminuria and gross proteinuria in persons with older-onset diabetes mellitus. Arch Intern Med 160:1093–1100
Fioretto P, Mauer M, Brocco E et al (1996) Patterns of renal injury in NIDDM patients with microalbuminuria. Diabetologia 39:1569–1576
Stevens LA, Coresh J, Greene T, Levey AS (2006) Assessing kidney function—measured and estimated glomerular filtration rate. N Engl J Med 354:2473–2483
Harjutsalo V, Katoh S, Sarti C, Tajima N, Tuomilehto J (2004) Population-based assessment of familial clustering of diabetic nephropathy in type 1 diabetes. Diabetes 53:2449–2454
Billings LK, Florez JC (2010) The genetics of type 2 diabetes: what have we learned from GWAS? Ann N Y Acad Sci 1212:59–77
Lindholm E, Agardh E, Tuomi T, Groop L, Agardh CD (2001) Classifying diabetes according to the new WHO clinical stages. Eur J Epidemiol 17:983–989
Ahluwalia TS, Lindholm E, Groop LC (2011) Common variants in CNDP1 and CNDP2, and risk of nephropathy in type 2 diabetes. Diabetologia 54:2295–2302
Alkhalaf A, Zurbig P, Bakker SJ et al (2010) Multicentric validation of proteomic biomarkers in urine specific for diabetic nephropathy. PLoS One 5:e13421
National Kidney Foundation (2002) K/DOQI clinical practice guidelines for chronic kidney disease: evaluation, classification, and stratification. Am J Kidney Dis 39: S1-266
Wong TY, Choi PC, Szeto CC et al (2002) Renal outcome in type 2 diabetic patients with or without coexisting nondiabetic nephropathies. Diabetes Care 25:900–905
Morris AD, Boyle DI, MacAlpine R et al (1997) The diabetes audit and research in Tayside Scotland (DARTS) study: electronic record linkage to create a diabetes register. DARTS/MEMO Collaboration. BMJ 315:524–528
Peterson B, Trell E, Kristenson H (1983) Comparison of gamma-glutamyltransferase and questionnaire test as alcohol indicators in different risk groups. Drug Alcohol Depend 11:279–286
Berglund G, Nilsson P, Eriksson KF et al (2000) Long-term outcome of the Malmo preventive project: mortality and cardiovascular morbidity. J Intern Med 247:19–29
Groop L, Forsblom C, Lehtovirta M et al (1996) Metabolic consequences of a family history of NIDDM (the Botnia study): evidence for sex-specific parental effects. Diabetes 45:1585–1593
Levey AS, Bosch JP, Lewis JB, Greene T, Rogers N, Roth D (1999) A more accurate method to estimate glomerular filtration rate from serum creatinine: a new prediction equation. Modification of Diet in Renal Disease Study Group. Ann Intern Med 130:461–470
Voight BF, Scott LJ, Steinthorsdottir V et al (2010) Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet 42:579–589
Dupuis J, Langenberg C, Prokopenko I et al (2010) New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet 42:105–116
Saxena R, Hivert MF, Langenberg C et al (2010) Genetic variation in GIPR influences the glucose and insulin responses to an oral glucose challenge. Nat Genet 42:142–148
Zeggini E, Scott LJ, Saxena R et al (2008) Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet 40:638–645
Yasuda K, Miyake K, Horikawa Y et al (2008) Variants in KCNQ1 are associated with susceptibility to type 2 diabetes mellitus. Nat Genet 40:1092–1097
Kottgen A, Pattaro C, Boger CA et al (2010) New loci associated with kidney function and chronic kidney disease. Nat Genet 42:376–384
McDonough CW, Palmer ND, Hicks PJ et al (2011) A genome-wide association study for diabetic nephropathy genes in African Americans. Kidney Int 79:563–572
Bianchi ME, Agresti A (2005) HMG proteins: dynamic players in gene regulation and differentiation. Curr Opin Genet Dev 15:496–506
Reeves R, Beckerbauer L (2001) HMGI/Y proteins: flexible regulators of transcription and chromatin structure. Biochim Biophys Acta 1519:13–29
Li O, Vasudevan D, Davey CA, Droge P (2006) High-level expression of DNA architectural factor HMGA2 and its association with nucleosomes in human embryonic stem cells. Genesis 44:523–529
Fusco A, Fedele M (2007) Roles of HMGA proteins in cancer. Nat Rev Cancer 7:899–910
Persson F, Andren Y, Winnes M et al (2009) High-resolution genomic profiling of adenomas and carcinomas of the salivary glands reveals amplification, rearrangement, and fusion of HMGA2. Gene Chromosome Canc 48:69–82
Fedele M, Battista S, Kenyon L et al (2002) Overexpression of the HMGA2 gene in transgenic mice leads to the onset of pituitary adenomas. Oncogene 21:3190–3198
Thuault S, Valcourt U, Petersen M, Manfioletti G, Heldin CH, Moustakas A (2006) Transforming growth factor-beta employs HMGA2 to elicit epithelial-mesenchymal transition. J Cell Biol 174:175–183
Acknowledgements
We would like to thank all the participants in the studies. We are very grateful to E. Lindholm (Department of Clinical Sciences, Diabetes and Endocrinology, Lund University, CRC, Skåne University Hospital, 205 02 Malmö, Sweden) for his contribution in the phenotyping of the SDR.
Funding
Studies at LUDC, Malmö, were supported by grants from the Swedish Research Council, including a Linné grant (No. 349-2008-6589), a strategic SFO grant, Exodiab (2009–1039), the Heart and Lung Foundation, the Swedish Diabetes Research Society, a European Community’s Seventh Framework Programme grant CEED3 (223211), the Diabetes Programme at the Lund University, the Påhlsson Foundation, the Crafoord Foundation, the Knut and Alice Wallenberg Foundation, Novo Nordisk Foundation and the European Foundation for the Study of Diabetes (EFSD). Steno received support from the EU Framework Programme 7 Syskid (FP7-241544). Go-DARTS received funding from a Wellcome Trust Bioresource grant and Scottish Health informatics grant (SHIP grant, 801969).
Duality of interest
The authors declare there is no duality interest associated with the manuscript.
Contribution statement
SA performed genotyping, participated in data analysis, interpretation, and manuscript writing. ML participated in data collection and manuscript writing. HD participated in data analysis and manuscript writing. HC participated in acquisition of data, conception and design of the Go-DARTS study. EA participated in data analysis of SDR. BI acquisition of data in the Botnia study. PR participated in acquisition of data, conception and design of the Steno study. LG participated in acquisition of data, conception and design of the Botnia study, participated in data interpretation. VL participated in the study design, data interpretation, manuscript writing and overall supervision of the study. All authors critically revised and approved the final version of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM Table 1
(PDF 17 kb)
ESM Table 2
(PDF 21 kb)
Rights and permissions
About this article
Cite this article
Alkayyali, S., Lajer, M., Deshmukh, H. et al. Common variant in the HMGA2 gene increases susceptibility to nephropathy in patients with type 2 diabetes. Diabetologia 56, 323–329 (2013). https://doi.org/10.1007/s00125-012-2760-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-012-2760-5