Abstract
Aims/hypothesis
Lon protease degrades oxidatively damaged proteins in mitochondrial matrix. To examine the relationships between mitochondrial quality control, mitochondrial functions and diabetes, we investigated whether lon protease deficiency influences insulin resistance by affecting mitochondrial function.
Methods
Lon protease-specific small interfering RNA (siRNA) was transfected into human liver SK-HEP-1 cells and changes in molecules related to insulin resistance were analysed.
Results
Reduction in lon protease was achieved using specific siRNA-induced mitochondrial dysfunction in human liver SK-HEP-1 cells. Concurrently, insulin signalling and subsequent insulin action were impaired and levels of gluconeogenic enzymes were increased by lon protein deficiency. Moreover, the activity of mitogen-activated protein kinases and transcription factors related to hepatic gluconeogenesis were elevated in LON (also known as LONP1) siRNA-transfected cells via increased intracellular reactive oxygen species production. Overproduction of lon protease restored mitochondrial function and also diminished the insulin resistance induced by treatment with cholesterol and palmitate. In addition, levels of lon protease decreased dramatically in livers of diabetic db/db mice compared with their lean mice counterparts.
Conclusions/interpretation
Here we have demonstrated that reduction of lon protease induced hepatic insulin resistance by lowering mitochondrial function. This is the first study to report that defects in mitochondrial protein quality control could cause insulin resistance and diabetes.
Similar content being viewed by others
Introduction
Accumulating research suggests that mitochondrial dysfunction is a cause of insulin resistance and type 2 diabetes [1, 2]. However, the mechanisms involved in the relationship between mitochondrial function and insulin resistance remain unclear. To fulfil their own functions, mitochondria contain approximately 700 proteins [3]. Three major mitochondrial proteases, lon, ClpP and AAA proteases are responsible for removal of misfolded or damaged proteins, thereby maintaining proper function of the mitochondria, similarly to the proteasome in the cytoplasm. While AAA proteases exist in the mitochondrial inner membrane, lon and ClpP are mitochondrial matrix proteins with lon being the most abundant protease [4].
Human lon protease, a homologue of bacterial La and yeast Pim1, degrades oxidatively damaged proteins [5]. This protease exists in most organisms and is highly conserved in structure, exhibiting a ring-shaped homo-oligomeric complex. Each monomer consists of three domains: (1) the substrate-binding N-terminal domain; (2) the ATPase domain (A-domain); and (3) the C-terminal proteolytically active domain (P-domain) [6]. The mechanism governing how lon protease recognises substrate proteins remains unclear, given that aconitase or Fe/S protein, the most well-known substrate of lon protease to date, contains no markers such as the ubiquitin tag. Substrate proteins are believed to be somehow recognised by lon protease via their exposed hydrophobic amino acid residues and may be translocated through the lon protease complex by hydrolysis of ATP, and then degraded [7, 8].
Lon protease is multifunctional. Aside from its proteolytic activity, it can bind to mDNA, particularly to regions located near the D-loop and to cytochrome c oxidase (COX) 1, and can also interact with mDNA polymerase γ as well as with the twinkle helicase. Therefore, lon protease may participate in replication/transcription of mDNA [9]. Additionally, lon protease exhibits chaperon-like functions that are independent of its proteolytic activity. Lon protease is also necessary for the assembly of mitochondrial electron transport chain complexes, especially COX II [10, 11]. Indeed, in the absence of Pim1, yeast becomes respiratory-deficient because it is unable to maintain mDNA, leading to accumulatin of electron-dense inclusion bodies in the mitochondrial matrix [12]. Finally, in humans, lon protease is associated with mitochondrial biogenesis; lon protease deficiency is responsible for the abnormal structure of mitochondria [13, 14].
We demonstrated here that reduction of lon protease production results in hepatic insulin resistance by inducing mitochondrial dysfunction. Lon protease deficiency appears to be correlated with pathological states, such as insulin resistance and diabetes.
Methods
Materials and animals
Oligomycin, cholesterol, palmitate, N-acetyl-cysteine (NAC) and the somatic cell ATP assay kit were purchased from Sigma (St Louis, MO, USA). The Bioxytech Aconitase-340 assay kit was obtained from Oxis Research (Foster City, CA, USA). Antibodies against phosphoenolpyruvate carboxykinase (PEPCK), peroxisome proliferator-activated receptor coactivator-1 alpha (PGC1α), glucose 6-phosphatase and myc were acquired from Santa Cruz Biotechnology (Santa Cruz, CA, USA). All other antibodies were purchased from Cell Signaling Technology (Beverly, MA, USA).
We obtained 10-week-old male db/db mice and age-matched lean mice (C57BL/6J) from the Jackson laboratory (Bar Harbor, ME, USA). After overnight fasting, mice were killed, and their livers removed and used for western blot analysis. All animal experiments were performed in compliance with the Korean Food and Drug Administration guidelines for care and use of laboratory animals.
Cell line and small interfering RNA of lon protease transfection
The SK-HEP-1 human liver cell line (ATCC HTB-52; ATTC, Manassas, VA, USA) was cultured in DMEM containing 10% (vol./vol.) fetal bovine serum and antibiotics.
Stealth LON (also known as LONP1)-specific small interfering RNA (siRNA) (oligo ID: HSS113887, HSS113888, HSS113889) and non-targeting control siRNA (12935-300) were purchased from Invitrogen (Carlsbad, CA, USA). Cells were transfected with 50 pmol of siRNA per 1.5 × 105 cells using lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. To confirm the reduction of LON expression, total RNA was extracted and reverse-transcribed using Super script II (Invitrogen) after 48 h of transfection, and PCR was then performed using primers as follows: forward AGATCATCAAGAAGGAGGAGGTGG, reverse CCGTCTTGGTCTTCTTCAAAC. In the event of stress induction by lipids, cells were treated with or without mixture containing 0.2 mmol/l cholesterol and 0.7 mmol/l palmitate at 24 h after transfection and incubated for a further 48 h.
Preparation of BSA-conjugated palmitate stock solution
Palmitate-BSA solution was prepared as described previously [15]. Palmitate was dissolved in 0.1 mmol/l NaOH at a concentration of 100 mmol/l by shaking before use, and incubated at 70°C. Palmitate stock solution (10 mmol/l) was prepared by adding 100 μl of 100 mmol/l palmitate dropwise to 900 μl of fatty acid-free BSA (10% wt/vol. in water; Sigma) and mixed at 55°C by shaking. Following a brief mixing, the palmitate-BSA solution was incubated for 10 min at 55°C and stored at −20°C after sterile filtration.
Measurement of 2-deoxy-d-[3H]glucose uptake
At 48 h after transfection and following serum starvation for 3 h, cells were washed twice with HEPES buffer (20 mmol/l HEPES [pH 7.4], 140 mmol/l NaCl, 2.5 mmol/l MgSO4, 5 mmol/l KCl, 1 mmol/l CaCl2) and pre-incubated with HEPES buffer for 1 h. After incubation for 10 min in the presence or absence of 200 nmol/l insulin, 2-deoxy-d-[3H]glucose (37,000 Bq/ml; Amersham Biosciences (Pittsburgh, PA, USA)) was applied to the cells for 10 min. The uptake was stopped by adding 10 μmol/l cytochalasin B. After washing with ice-cold 0.9% (wt/vol.) NaCl, total cells were lysed by 0.1 mol/l NaOH. Non-specific uptake was measured in the presence of 10 μmol/l cytochalasin B and was subtracted from all the values.
Measurement of reactive oxygen species
Formation of intracellular reactive oxygen species (ROS) was monitored using 5-(and-6-)-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate, acetyl ester (CM-H2DCFDA; Molecular Probes, Eugene, OR, USA). After transfection of siRNA of lon protease or treatment with the mixture of cholesterol and palmitate (0.2 mmol/l cholesterol, 0.7 mmol/l palmitate), cells were incubated for 30 min with 5 μmol/l CM-H2DCFDA in the presence of medium without phenol red. After washing with PBS, fluorescence produced from 2′,7′-dichlorodihydrofluorescein, which represented the oxidised product, was detected by confocal microscopy at excitation 490 nm and emission 535 nm (Radiance 2000; Bio-Rad, Hercules, CA, USA).
Construction of the lon protease expression vector and generation of lon protease antibody
The human lon protease expression vector and antibody were manufactured using custom-built services of COSMO Gene Technology (Seoul, South Korea). Briefly, to obtain the pLon-myc expression vector, a 3,137-bp fragment of full-length human lon protease (NCBI accession number: BC000235) was amplified by PCR using the following primers: forward,CA GGATCC ACATGGCGGCGAGCACTGGCTA, reverse CGGAATTC CCGTTCCACGGCCAGCGCCTC, and cloned into the vector (pcDNA 3.1/myc-HisA vector; Invitrogen).
To manufacture the antibody specific for lon protease, a peptide corresponding to the amino acids from positions 410–427 of lon protease, LEKDDKDAIEEKFRERLKE, was synthesised and an extra C introduced at the N-terminus of the peptide to conjugate the keyhole limpet haemocyanin. The product was used to immunise rabbits. Following the third immnunisation, antibodies were purified and stored at −20°C.
Determination of lon protease localisation
Lon protease localisation was checked by immunohistochemistry. Cells were transfected either with pLon-myc or pcDNA3.1 using a transfection reagent (Lipofectamine; Invitrogen). After 48 h of transfection, cells were fixed with 100% ice-cold methanol and incubated for 1 h with 10% (vol./vol.) goat serum in PBS. Cells were washed carefully with PBS and incubated for 1 h, either with anti-goat COX1 or anti-mouse myc antibodies in 10% (vol./vol.) goat serum/PBS. After incubation of a second antibody for goat (Alexa522) and mouse (Alexa488), fluorescence signals of lon protease compared with COX1 were detected by confocal microscopy (Radiance 2000; Bio-Rad).
Statistical analysis
All results are expressed as the mean ± SE. Statistical analysis was performed using Student’s t test and data were considered statistically significant at p < 0.05.
Results
Reduction of lon protease induces mitochondrial dysfunction
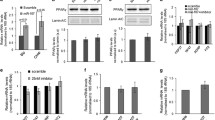
Mitochondrial lon protease is responsible for removing oxidatively damaged proteins from the mitochondria. To investigate whether a decline in lon protease expression affected mitochondrial function in our system, three different LON-specific siRNAs (Loni) were transfected into human liver SK-HEP-1 cells and the degree of lon protease reduction was measured by RT-PCR. Of these three, only one (oligoID: HSS113888) decreased the expression of lon protease and this Loni was used for further experiments (Fig. 1a). The results of the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazoliumbromide (MTT) assay demonstrated that transfection with Loni did not alter cell viability of SK-HEP-1 cells (Fig. 1c). However, aconitase activity was reduced by approximately 30% in cells transfected with Loni compared with cells transfected with the negative control siRNA (Conti) (Fig. 1b). Interestingly, total cellular ATP contents and mitochondrial membrane potential (ΔΨm) were significantly reduced by reduction of lon protease (Fig. 1d, e). These results confirmed that lon protease deficiency lowers mitochondrial function.
Lon protease deficiency induces mitochondrial dysfunction a Reduction of lon protease expression by transfection with LON-siRNA. RT-PCR was performed as described in the Methods. b Reduction of aconitase activity in SK-HEP-1 human liver cells. After transfection of siRNA as indicated, aconitase activity was measured according to the manufacturer’s instructions (Oxis Research). The aconitase activity from control siRNA-transfected cells was set to 1; n = 3, *p < 0.05. c Determination of cell viability using 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl tetrazolium bromide (MTT). Cells were treated as indicated, the formazan products were solubilised by dimethulsulfoxide and absorbance was measured at 570 nm. Values for the control were set to 1. d Measurement of the ATP content of LON siRNA-transfected cells relative to the control siRNA in SK-HEP-1 cells as described previously [16]. The ATP content from control siRNA-transfected cells was set to 100%; n = 3, ***p < 0.001. e Reduction of mitochondrial membrane potential. Cells stained with 100 nmol/l MitoTracker Red FM were visualised by confocal microscopy at a magnification of ×600. Images were pseudo-coloured (red) according to the fluorescence intensity
Lon protease deficiency causes abnormally increased hepatic gluconeogenesis
In previous reports, we have shown that mitochondrial dysfunction induces hepatic insulin resistance via activation of p38 mitogen-activated protein kinase (MAPK) and its downstream transcription factors [16]. Given that a reduction of lon protease induces mitochondrial dysfunction, we hypothesised that lon protease deficiency would cause hepatic gluconeogenesis to increase. In addition to transfection of Loni, cells were treated with a mixture of cholesterol and palmitate, a known inducer of hepatic insulin resistance. As shown in Fig. 2a, levels of PEPCK, the key enzyme of gluconeogenesis, were greatly increased by transfection with Loni alone and to an even greater degree than by cholesterol and palmitate treatment. In addition, levels of glucose-6-phosphatase and PGC-1α were significantly increased by reduction of lon protease, suggesting that lon protease deficiency is responsible for the abnormally increased hepatic gluconeogenesis.
Induction of hepatic insulin resistance by reduction of lon protease expression. At 48 h after transfection with Loni or treatment with cholesterol and palmitate (CP) (0.2 mmol/l and 0.7 mmol/l respectively), cells were collected and subjected to western blot analysis. a Changes in protein levels associated with gluconeogenesis, (b) ER stress markers and (c) and lon protease were determined. G6Pase, glucose 6-phosphatase; eIF2α, eukaryotic initiation factor 2 α; GRP78, glucose-regulated protein 78 kDa
Endoplasmic reticulum (ER) stress is considered to be a critical component in the development of insulin resistance and type 2 diabetes [17]. To ascertain whether silencing of lon protease also induces ER stress, thereby increased hepatic gluconeogenesis, we examined the changes of ER stress markers after transfection with Loni. Figure 2b shows that reduction of lon protease expression did not induce ER stress. In contrast, treatment with cholesterol and palmitate, known inducers of ER stress, did trigger ER stress [18, 19] (Fig. 2b). Hypoxia and ER stress increase expression of lon protease [10]. Since cholesterol and palmitate triggered ER stress, the influences of this treatment on expression of lon protease were investigated. The level of lon protease was increased by cholesterol and palmitate. However, transfection with Loni was able to effectively block the elevation of lon production even in the presence of cholesterol and palmitate (Fig. 2c).
To gain further insight into how hepatic gluconeogenesis is elevated in cells with Loni, the changes in factors related to gluconeogenesis were investigated. Several MAPKs are known as stress-inducible kinases that are also induced by mitochondrial dysfunction. As shown in Fig. 3a, basal phosphorylation of c-Jun N-terminal kinase (JNK) and p38 MAPK was elevated by lon protease deficiency, although total levels of these protein kinases remained unchanged. In contrast, neither the protein level nor the phosphorylation of extracellular signal-regulated kinase 1/2 was affected by changes in lon protease expression.
Abnormal increase of stress kinases and transcription factors related to hepatic gluconeogenesis. Treatment with cholesterol/palmitate (CP) was done as described in Methods. Whole-cell lysates were obtained after treatment of cells as indicated and western blot analysis performed using antibodies for MAPKs (a) or transcription factors (b). c The intensity of phosphorylated (p):whole protein levels of c/EBPα and (d) ATF2, relative to the values for non-treated cells, as indicated. Values represent the mean±SE from three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001
Of the numerous transcription factors that regulate production of PEPCK, PGC-1α, activating transcription factor (ATF)2 and CCAAT/enhancer-binding protein (c/EBP)α participate in upregulation of PEPCK under the control of p38 MAPK. We therefore tested whether production or phosphorylation of these factors was altered by transfection with Loni. We found that, in addition to elevation of PGC-1α levels (Fig. 2a), phosphorylation of c/EBPα and ATF2 was increased in cells transfected with Loni (Fig. 3b–d).
Disturbance of insulin action due to reduction of lon protease expression
Hepatic gluconeogenesis is controlled by insulin. To determine whether the reduction of lon protease hindered insulin action, thereby causing abnormal elevation of hepatic gluconeogenesis in Loni transfected cells, we first investigated whether insulin signalling was affected by lon protease deficiency. Figure 4a shows that serine phosphorylation of Akt by insulin was diminished by the transfection with Loni, implying that a reduction in lon protease expression disrupts insulin signalling. Next we examined whether impaired insulin signalling by deficiency of lon protease attenuated insulin-mediated repression of PEPCK (also known as PCK1) expression. Compared with Conti-transfected cells, where insulin treatment caused significant reduction of PEPCK, Loni-transfected cells did not respond to insulin and PEPCK expression remained unchanged by insulin treatment (Fig. 4b). To ascertain whether lon deficiency caused insulin insensitivity of liver cells, we also measured glucose uptake, since insulin not only inhibits gluconeogenesis, but also stimulates glucose uptake in liver to maintain glucose homeostasis in the whole body [20, 21]. Whereas insulin-stimulated glucose uptake was 2.7-fold increased in Conti-treated cells, it was not observed in Loni-transfected cells (Fig. 4c).
Impairment of insulin action in Loni-transfected cells. a Serine phosphorylation of Akt (pAkt) protein was determined by western blot analysis, followed by application of insulin (200 nmol/l) for 10 min after 3 h serum starvation. The intensity of phosphorylated:whole protein levels was quantified relative to the values for non insulin-treated cells. b Disturbance of insulin action by reduction of lon protease expression. At 48 h after transfection with Loni, cells were treated with insulin (200 nmol/l) for 4 h after serum starvation. Cells were collected and subjected to RT-PCR analysis. Changes in the expression of PEPCK by insulin treatment were measured by RT-PCR using gene-specific primers (forward, ATCGAAAGCAAGACGGTTAT, reverse, CAATGCCTTCCCAGTAAAC). c Insulin-stimulated uptake of 2-deoxy-d-[3H]glucose (2-DG) was measured as described in Methods. Values for non insulin-treated cells were set to 1 and are represented as relative values. d Increased accumulation of triacylglycerol (TG) was quantified colorimetrically as described previously [34]. All experimental results are presented as mean±SE; n = 3; *p < 0.05; **p <0.01; ***p < 0.001
An accumulation of lipids, especially triacylglycerol, in cells leads to the activation serine/threonine kinases, which interfere with insulin signalling by increasing phosphorylation of IRS-1 at the specific serine residues [22, 23]. To elucidate how deficiency of lon protease could disturb insulin action, we measured triacylglycerol accumulation in Loni-transfected cells. We found that the cellular triacylglycerol contents were elevated in the Loni-transfected cells and that cholesterol and palmitate treatment of Loni-transfected cells further increased triacylglycerol accumulation (Fig. 4d). From these results, we postulated that reduction of lon protease impaired insulin signalling and insulin action, thereby inducing an abnormal increase of gluconeogenesis and insulin resistance in liver cells.
Increase of ROS by lon protease deficiency causes abnormal elevation of gluconeogenesis
Mitochondria are major sources for the generation of ROS, and mitochondrial dysfunction contributes to greater ROS generation. We showed that lon protease deficiency is a cause of mitochondrial dysfunction. Therefore, we postulated that an increase in hepatic gluconeogenesis may be due to elevation of ROS generation through the reduction of lon protease expression. First, we examined whether lon protease deficiency had any influence on the generation of ROS. Figure 5a shows that transfection with Loni alone significantly increased the amount of ROS compared with transfection with Conti to an even greater degree than the generation of ROS by the cholesterol and palmitate treatment. To determine whether elevation of ROS by reduced lon protease expression was involved in the abnormal increase of hepatic gluconeogenesis, NAC, an antioxidant, was administrated during transfection with Loni and changes in the abundance of factors related to gluconeogenesis were examined. Similar to the observation that cholesterol and palmitate treatment-induced production of PEPCK was reduced by NAC treatment, NAC effectively blocked the overproduction of PEPCK by lon protease deficiency (Fig. 5b). In addition, cholesterol- and palmitate treatment or lon protease deficiency induced PGC-1α overproduction and p38 MAPK phosphorylation (pp38), factors that are responsible for increased gluconeogenesis, were reduced by co-treatment with NAC, suggesting that elevation of ROS by lon protease deficiency in liver cells is somehow related to an abnormal increase in hepatic gluconeogenesis.
Increased intracellular ROS formation is responsible for the elevation of gluconeogenesis by reduction of lon protease expression. a ROS levels were measured using CM-H2DCFDA as described in the Methods. Pseudo-coloured (green) fluorescence signals were visualised by confocal microscopy at a magnification of ×100. b Cells were transfected as indicated. NAC (15 mmol/l) was applied for 30 min, before addition of cholesterol/palmitate (CP), after which cells were further incubated for 48 h. Western blots were performed to detect changes in proteins related to gluconeogenesis
Overexpression of lon protease ameliorates hepatic insulin resistance
To elucidate whether a reduction in lon protease directly causes mitochondrial dysfunction and concomitant induction of hepatic insulin resistance, we introduced a myc-tagged lon protease expression vector (pLon-myc) to cells containing dysfunctional mitochondria. First, we tested whether lon protease was expressed properly in the mitochondria. We used COX1 as a marker for mitochondria. The localisation of lon protease was detected using an anti-myc antibody. As shown in Fig. 6a, an anti-myc antibody signal was detected in cells transfected with pLon-myc, but not in cells with an empty pcDNA3.1 vector. The co-presence of anti-myc and anti-COX1 signals indicated that expressed lon protease was properly targeted in the mitochondria.
Amelioration of insulin resistance by overexpression of lon protease. a Localisation of expressed lon protease. Lon and COX1 were visualized with Alexa 488 (green) and Alexa 522 (red)-conjugated secondary antibodies, respectively; magnification ×600. b Recovery of cellular ATP contents. Cholesterol and palmitate (CP) were introduced 24 h after transfection with pcDNA3.1 or pLon-myc. Cells were incubated for another 48 h and cellular ATP content was measured on basis of ATP-driven luciferin luciferase activity; n = 3; *p < 0.05; ***p < 0.001. c, d Western blot analysis followed by overexpression of pLon-myc in the presence of cholesterol and palmitate using antibodies as indicated. Erk, extracellular signal-regulated kinase
Next, we tested whether overexpression of lon protease could restore mitochondrial function after it had been reduced by cholesterol and palmitate treatment. Mitochondrial function was estimated by measuring the total cellular ATP content. Figure 6b shows that the reduction of total ATP by lipid treatment was effectively restored by overproduction of lon protease. In turn, we investigated whether restoration of mitochondrial function by lon protease expression could diminish overproduction of PEPCK. Following transfection either with pLon-myc or empty pcDNA3.1 vector, cells were treated with cholesterol and palmitate for 48 h and western blotting performed. The elevated levels of PEPCK and PGC-1α in lipid-treated cells were reduced concurrently with the recovery of mitochondrial function by lon protease expression (Fig. 6c). In addition, the activities of p38 MAPK and JNK were reduced by lon protease expression as in control cells, implying that lon protease was able to relieve stress responses that cause mitochondrial dysfunction and concomitant insulin resistance (Fig. 6d).
Changes in expression of lon protease in diabetic murine liver
Increasing evidence supports a correlation between mitochondrial dysfunction and diabetes. Given that lon protease is a major factor in maintaining mitochondrial function, we postulated that lon protease levels might decrease under diabetic conditions, which are often related to mitochondrial dysfunction. To determine lon protease protein levels in mice, we generated an antibody specific to lon protease and obtained an approximately 100 kDa lon protease immunoreactive band (data not shown). Whole proteins were extracted from the liver of db/db mice and their age-matched lean mice, and changes in lon protease production were examined. While the level of cytosolic heat shock protein (HSP) 70, a stress-inducible chaperone, remained unchanged (Fig. 7a, c), the protein level of lon protease was drastically reduced in the livers of db/db mice (Fig. 7a, b), a finding possibly associated with the reduction in mitochondrial function and concomitant increase of hepatic gluconeogenesis in db/db mice.
Reduced expression of lon protease in diabetic mice. The livers were extracted from 10-week-old male db/db and age-matched lean mice and western blotting (a) performed. Protein levels obtained by densitometry (b, c) were normalised against tubulin signals. The intensity for control was set to 100%. ***p < 0.001
Discussion
Several conditions, including insulin resistance and diabetes, are closely related to the ER stress response, also known as the unfolded protein response (UPR), which is triggered by an accumulation of unfolded proteins in the ER lumen [24, 25]. Similar to the UPR in the ER, a mitochondria-specific stress response, termed mitochondrial UPR, was recently reported. With an accumulation of unfolded, damaged proteins in the mitochondrial matrix, the levels of mitochondrial chaperones and proteases such as mitochondrial HSP70, mitochondrial HSP60 and the mitochondrial homologue of the bacterial ClpP protease (mtClpP) were upregulated [26, 27]. However, unlike the UPR in the ER, the molecular mechanisms of the mitochondrial UPR and the correlation between the mitochondrial UPR and disease remain to be identified. Here, we showed that a decrease in mitochondrial lon protease, one of the components of mitochondrial protein quality control, leads to hepatic insulin resistance by disturbing insulin action and to concomitant elevation of gluconeogenesis.
Given that lon protease is a major factor in the removal of oxidatively damaged aconitase and the assembly of mitochondrial electron transport components such as COX, its reduction by transfection with of Loni caused mitochondrial dysfunction (Fig. 1) and concomitant elevation of ROS generation (Fig. 5a). We postulated that defects in the mitochondrial electron transport system induced by lon protease deficiency may cause irregular side reactions that lead to ROS generation. ROS are known to activate stress-activated kinases such as p38 MAPK and JNK, as well as disturbing insulin signalling by increasing serine phosphorylation of IRS-1 at specific sites [28]. Indeed, we found that lon protease deficiency resulted in activation of p38 MAPK and JNK, and also disturbed insulin signalling (Figs 3a, and 4a) and activation of transcription factors related to gluconeogenesis by p38 MAPK (Fig. 3b, c, d), which together are responsible for the abnormal elevation of hepatic gluconeogenesis. In addition to the disturbance in insulin signalling, we showed that insulin-mediated repression of PEPCK expression and insulin-stimulated glucose uptake disappeared upon reduction of lon protease expression (Fig. 4b, c). Together, this implies that deficiency of lon protease could trigger hepatic insulin resistance by elevation of gluconeogenesis and impairment of insulin signalling and action in the liver cells.
Elevation of ROS generation by lon protease deficiency may lead to increased oxidatively damaged proteins in cells; however, cells cannot remove these damaged proteins effectively due to reduced production of lon protease. This may cause further disturbances to mitochondrial function, leading to subsequent worsening of the stress response. In addition, in cells transfected with Loni, we showed intracellular accumulation of triacylglycerol, which also contributes to activation of stress kinases and repression of insulin signalling by phosphorylation of IRS-1 (Fig. 4d) [22]. We postulate that elevated ROS generation and triacylglycerol accumulation together create a vicious circle that leads to hepatic insulin resistance. Based on our results, we also hypothesise that mitochondrial lon protease in normal conditions can maintain fitness of mitochondria and protect them against stressors that might trigger insulin resistance. However, when these stressors somehow exceed the capacity of lon protease or repress its production, mitochondria become dysfunctional, ultimately resulting in insulin resistance. Overproduction of lon protease in liver cells effectively prevented insulin resistance induced by cholesterol and palmitate treatment (Fig. 6). Moreover, the protein level of lon protease was severely reduced in the livers of db/db mice (Fig. 7), supporting our hypothesis. Indeed, lon protease production and activity diminish with age, when mitochondria show increased dysfunction and subsequent elevation of tendency to develop insulin resistance [6, 11, 29–31]. Similar to the situation in old mice, mitochondrial function should be decreased in db/db mice. Mitochondrial lon protease, which is translated in the cytosol, should be imported into mitochondria by mitochondrial membrane potential and ATP [4, 7]. Therefore, in db/db mice, which have more dysfunctional mitochondria, the import of lon protease is somehow interfered, possibly reducing the amount of functionally active lon protease in the mitochondria. The deficiency of functionally active lon protease could cause further mitochondrial dysfunction. We hypothesised that reduction of lon protease expression or impairment of lon activity plays an important role in triggering abnormality of insulin action by disturbance of mitochondrial function. As lon deficiency continues, increased mitochondrial dysfunction might act to further stimulate reduction of lon protease activity or expression, creating a vicious cycle that ultimately leads to insulin resistance and type 2 diabetes.
Recently, Civitarese et al. reported that depletion of presenillin-associated rhomboid-like (PARL), a yeast mitochondrial protease purkinje cell protein 1 homologue, causes type 2 diabetes by inducing mitochondrial dysfunction. They showed that knockdown of PARL in muscle leads to mitochondrial dysfunction and subsequent impairment of insulin signalling through increased ROS generation; they also showed that the abundance of PARL was reduced in elderly and type 2 diabetes patients [32]. PARL is known as a mitochondrial processing peptidase located in mitochondrial inner membrane [7]. Even though the molecular mechanisms behind mitochondrial proteases and mitochondrial URP remain elusive, it appears that mitochondrial proteases such as PARL and lon protease are closely related to triggering of insulin resistance.
Lon protease expression is stress-inducible; however, little is known about the regulation of lon protease expression. Hypoxia and ER stress are known to stimulate lon protease expression [10]. Here, treatment with cholesterol and palmitate induced ER stress and concomitant stimulation of lon protease expression (Fig. 2b, c). We assume that increase of lon protease expression under cholesterol and palmitate treatment is a protective mechanism of cells to maintain mitochondrial function against stressor, similar to the induction of glucose-regulated protein 78 kDa under ER stress responses [17, 24]. However, silencing of lon protease effectively blocked the increase of lon protease expression by treatment with cholesterol and palmitate (Fig. 2c). Even though treatment with cholesterol and palmitate, and transfection with Loni finally caused hepatic gluconeogenesis to increase and induced insulin resistance, the process that leads to insulin resistance does not seem to be identical.
Bota et al. have shown that downregulation of lon protease in WI-38 VA-13 human lung fibroblast leads to mitochondrial dysfunction and apoptosis [14]. We observed mitochondrial dysfunction, but did not see cell death by transfection with Loni (Fig. 1c). We postulate that the different cells used in the experiments (human lung fibroblasts vs human liver cells) and the degree of reduction of lon protease expression may explain this discrepancy. Bota et al. also described that the expression of lon protease was not detected 4 days after transfection with lon antisense oligonucleotide. Conversely, we found reduced but still detectable expression of lon protease at 3–4 days after transfection with Loni (Fig. 1a). Similarly to the way ER stress, which induces insulin resistance, can cause apoptosis when stress exceeds a threshold, the reduction of lon protease and concomitant mitochondrial dysfunction may at first lead to insulin resistance, then gradually cause cellular apoptosis by increasing the degree of lon protease reduction. Indeed, overproduction of lon protease causes death in Escherichia coli and leads to tumorigenic transformation in eukaryotes [33]; moreover, reduced production of lon protease leads to cellular defects as shown by others [5, 9] and our results here, implying that expression of lon protease must be tightly controlled. However, the mechanisms or factors participating in the regulation of lon protease remain to be established.
In summary, we demonstrated a unique molecular mechanism that causes hepatic insulin resistance. Specifically, lon protease deficiency leads to mitochondrial dysfunction, which ultimately leads to insulin resistance. Thus, mitochondrial lon protease acts to maintain the fitness of mitochondria, thereby preventing conversion to a pathological status.
Abbreviations
- ATF:
-
Activating transcription factor 2
- c/EBP:
-
CCAAT/enhancer-binding protein
- CM-H2DCFDA:
-
5-(and-6-)-Chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate, acetyl ester
- Conti:
-
Control siRNA
- COX:
-
Cytochrome c oxidase
- ER:
-
Endoplasmic reticulum
- HSP:
-
Heat shock protein
- JNK:
-
c-Jun N-terminal kinase
- Loni:
-
LON-specific siRNA
- MAPK:
-
Mitogen-activated protein kinase
- NAC:
-
N-Acetyl-cysteine
- PARL:
-
Presenillin-associated rhomboid-like
- PEPCK:
-
Phosphoenolpyruvate carboxykinase
- PGC-1α:
-
Peroxisome proliferator-activated receptor coactivator-1 alpha
- ROS:
-
Reactive oxygen species
- siRNA:
-
Small interfering RNA
- UPR:
-
Unfolded protein response
References
Lowell BB, Shulman GI (2005) Mitochondrial dysfunction and type 2 diabetes. Science 307:384–387
Johannsen DL, Ravussin E (2009) The role of mitochondria in health and disease. Curr Opin Pharmacol 9:780–786
Prokisch H, Scharfe C, Camp DG et al (2004) Integrative analysis of the mitochondrial proteome in yeast. PLoS Biol 2:795–804
Voos W (2009) Mitochondrial protein homeostasis: the cooperative roles of chaperones and proteases. Res Microbiol 160:718–725
Bota DA, Davies KJA (2002) Lon protease preferentially degrades oxidized mitochondrial aconitase by an ATP-stimulated mechanism. Nat Cell Biol 4:674–680
Bulteau A-L, Szweda L, Friguet B (2006) Mitochondrial protein oxidation and degradation in response to oxidative stress and aging. Exp Gerontol 41:653–657
Koppen M, Langer T (2007) Protein degradation within mitochondria: versatile activities of AAA proteases and other peptidases. Crit Rev Biochem Mol Biol 42:221–242
Ondrovicova G, Liu T, Singh K et al (2005) Cleavage site selection within a folded substrate by the ATP-dependent lon protease. J Biol Chem 280:25103–25110
Lu B, Yadav S, Shah PG, Liu T et al (2007) Roles for the human ATP-dependent lon protease in mitochondrial DNA maintenance. J Biol Chem 282:17363–17374
Hori O, Ichinoda F, Tamatani T et al (2002) Transmission of cell stress from endoplasmic reticulum to mitochondria: enhanced expression of lon protease. J Cell Biol 157:1151–1160
Ngo JK, Davies KJA (2007) Importance of the lon protease in mitochondrial maintenance and the significance of declining lon in aging. Ann NY Acad Sci 1119:78–87
Suzuki CK, Suda K, Wang N, Schatz G (1994) Requirement for the yeast gene lon in intramitochondrial proteolysis and maintenance of respiration. Science 264:273–276
Lucaikova K, Sokolokova B, Chloupkova M, Nelson BD (1999) Enhanced mitochondrial biogenesis is associated with increase expression of the mitochondrial ATP-dependent lon protease. FEBS Lett 444:186–188
Bota DA, Ngo JK, Davies KJA (2005) Downregulation of the human lon protease impairs mitochondrial structure and function and causes cell death. Free Radic Biol Med 38:665–677
Cousin SP, Huegl SR, Wrede CE, Kajio H, Myers MG Jr, Rhodes CJ (2001) Free fatty acid-induced inhibition of glucose and insulin-like growth factor I-induced deoxyribonucleic acid synthesis in the pancreatic b-cell line INS-1. Endocrinology 142:229–240
Lim JH, Lee HJ, Jung MH, Song J (2009) Coupling of mitochondrial dysfunction to endoplasmic reticulum stress response: a molecular mechanism leading to hepatic insulin resistance. Cell Signal 21:169–177
Özcan U, Yilmaz E, Lee AH et al (2004) Endoplasmic reticulum stress links obesity, insulin action, and type2 diabetes. Science 306:457–561
Mayer CM, Belsham DD (2010) Palmitate attenuates insulin signaling and induces endoplasmic reticulum stress and apoptosis in hypothalamic neurons: rescue of resistance and apoptosis through adenosine 5′monophosphate-activated protein kinase activation. Endocrinology 151:576–585
Pineau L, Colas J, Dupont S et al (2009) Lipid-induced ER stress; synergistic effects of sterols and saturated fatty acids. Traffic 10:673–690
Iozzo P, Geisler F, Oikonen V et al (2003) Insulin stimulates liver glucose uptake in humans: an 18F-FDG PET study. J Nucl Med 44:682–689
Cherrington AD (1999) Control of glucose uptake and release by the liver in vivo. Diabetes 48:1198–1214
Vankoningsloo S, Piens M, Lecocq C et al (2005) Mitochondrial dysfunction induces triglyceride accumulation in 3T3-L1 cells: role of fatty acid beta-oxidation and glucose. J Lipid Res 46:1133–1149
Petersen KF, Dufour S, Befroy D, Garcia R, Shulman GI (2004) Impaired mitochondrial activity in the insulin-resistant offspring of patients with type 2 diabetes. N Engl J Med 350:664–671
Özcan U, Yilmaz E, Özcan L et al (2006) Chemical chaperones reduce ER stress and restore glucose homeostasis in a mouse model of type 2 diabetes. Science 313:1137–1140
Laybutt DR, Preston AM, Akerfeldt MC et al (2007) Endoplasmic reticulum stress contributes to beta cell apoptosis in type 2 diabetes. Diabetologia 50:752–763
Zhao Q, Wang J, Levichkin IV, Stasinopoulos S, Ryan MT, Hoogenraad J (2002) A mitochondrial specific stress response in mammalian cells. EMBO J 21:4411–4419
Figuet B, Bulteau AL, Petropoulos I (2008) Mitochondrial protein quality control: implications in ageing. Biotechnol J 3:757–764
Newsholme P, Haber EP, Hirabara SM et al (2007) Diabetes associated cell stress and dysfunction: role of mitochondrial and non-mitochondrial ROS production and activity. J Physiol 583:9–24
Bota DA, Remmen HV, Davies KJA (2002) Modulation of lon protease activity and aconotase turnover during aging and oxidative stress. FEBS Lett 532:103–106
Lee CK, Klopp RG, Weindruch R, Prolla TA (1999) Gene expression profile of aging and its retardation by caloric restriction. Science 285:1390–1393
Bakala H, Delaval E, Hamelin M et al (2003) Changes in rat liver mitochondria with aging. Lon protease-like reactivity and N(epsilon)-carboxymethyllysine accumulation in the matrix. Eur J Biochem 270:2295–2302
Civitarese AE, Maclean PS, Carling S et al (2010) Regulation of skeletal muscle oxidative capacity and insulin signaling by the mitochondrial rhomboid protease PARL. Cell Metab 11:412–426
Christensen SK, Maenhaut-Michel G, Mine N, Gottesman S, Gerdes K, van Melderen L (2004) Overproduction of the lon protease triggers inhibition of translation in E. coli. Mol Miocrobiol 51:1705–1717
Lim JH, Lee JI, Suh YH, Kim W, Song J, Jung MH (2006) Mitochondrial dysfunction induces aberrant insulin signaling and glucose utilisation in murine C2C12 myotube cells. Diabetologia 49:1924–1936
Acknowledgements
This study was supported by research grants from the Korean National Institute of Health (4845-300-210-13).
Duality of interest
The authors declare that there is no duality of interest associated with this manuscript.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Lee, H.J., Chung, K., Lee, H. et al. Downregulation of mitochondrial lon protease impairs mitochondrial function and causes hepatic insulin resistance in human liver SK-HEP-1 cells. Diabetologia 54, 1437–1446 (2011). https://doi.org/10.1007/s00125-011-2074-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-011-2074-z