Abstract
Aims/hypothesis
The basic helix–loop–helix transcription factor neurogenin-3 (NGN3) commits the fates of pancreatic progenitors to endocrine cell types, but knowledge of the mechanisms regulating the choice between proliferation and differentiation of these progenitors is limited.
Methods
Using a chromatin immunoprecipitation cloning approach, we searched for direct targets of NGN3 and identified a zinc-finger transcription factor, OVO homologue-like 1 (OVOL1). Transactivation experiments were carried out to elucidate the functional role of NGN3 in Ovol1 gene expression. Embryonic and adult rodents pancreases were immunostained for OVOL1, Ki67 and NGN3.
Results
We showed that NGN3 negatively regulates transcription of Ovol1 in an E-box-dependent fashion. The presence of either NGN3 or NEUROD1, but not MYOD, reduced endogenous Ovol1 mRNA. OVOL1 was detected in pancreatic tissue around embryonic day 15.5, after which OVOL1 levels dramatically increased. In embryonic pancreas, OVOL1 protein levels were low in NGN3+ or Ki67+ cells, but high in quiescent differentiated cells. OVOL1 presence was maintained in adult pancreas, where it was detected in islets, pancreatic ducts and some acinar cells. Additionally OVOL1 presence was lacking in proliferating ductules in regenerating pancreas and induced in cells as they began to acquire their differentiated phenotype.
Conclusions/interpretation
The timing of OVOL1 appearance in pancreas and its increased levels in differentiated cells suggest that OVOL1 promotes the transition of cells from a proliferating, less-differentiated state to a quiescent more-differentiated state. We conclude that OVOL1, a downstream target of NGN3, may play an important role in regulating the balance between proliferation and differentiation of pancreatic cells.
Similar content being viewed by others
Introduction
There is a critical need to develop a reliable source of beta cells for the treatment of diabetes. To accomplish this, it is necessary to understand the process regulating beta cell differentiation. Knockout and transgenic studies demonstrate that Ngn3 (also known as Neurog3) is essential and sufficient to initiate pancreatic endocrine differentiation [1–4]. Ngn3-expressing cells can be detected as early as embryonic day (E) 9.5, remain few in number until E12.5 and peak at E14.5, rapidly decreasing to undetectable levels after birth [5]. Such transient expression of the gene encoding neurogenin-3 (NGN3) is sufficient to induce expression of the genes encoding several transcription factors, e.g. NK2 transcription factor related, locus 2 (NKX2.2), paired box gene 4 (PAX4) and neurogenic differentiation 1 (NEUROD1), that regulate endocrine differentiation [6–8]. NGN3+ progenitors have a very limited capacity to proliferate [9–11]. Our understanding of the factors regulating the transient expression of Ngn3 and the capacity of endocrine progenitors to proliferate is limited. Approaches based on large-scale analysis of gene expression have successfully identified several downstream targets of NGN3 [12–14]. However, one of the limitations of such analyses is the difficulty in distinguishing direct vs indirect targets of NGN3. To identify direct targets of NGN3, we used a chromatin immunoprecipitation (ChIP)-based cloning approach. This approach identified a zinc-finger transcription factor OVO homologue-like 1 (OVOL1), which in skin epidermis promotes the transition of precursor cells from a proliferating, less differentiated state to a post-mitotic more differentiated state [15]. We show that OVOL1 is a direct target of NGN3 and that, unlike its other targets such as NEUROD1, NKX2.2, PAX4, insulinoma-associated 1 and myelin transcription factor 1, which are activated by NGN3 [6, 8, 16], Ovol1 expression is inhibited by NGN3. Consistent with this observation, cells showing high levels of NGN3 have reduced levels of OVOL1; high OVOL1 level was observed in quiescent cells. We suggest that OVOL1 may regulate the capacity of endocrine progenitors to proliferate. Inhibiting its function, therefore, could enhance the proliferation of endocrine precursors resulting in a greater yield of beta cells.
Methods
Cell lines and plasmids
AR42J-B13 (rat pancreatic cell line provided by I. Kojima, Institute for Molecular and Cellular Regulation, Gunma University, Gunma, Japan), INS-1 (rat insulinoma cell line provided by C. B. Wollheim, Department of Cell Physiology and Metabolism, University of Geneva, Geneva, Switzerland) and 293Ad cells (Stratagene, La Jolla, CA, USA) were cultured as previously described [17–19]. Ngn3 (gift of D. J. Anderson, Howard Hughes Medical Institute, Pasadena, CA, USA) and Neurod1 [20] complete DNA coding sequences were amplified by PCR and subcloned into mammalian expression vector pcDNA5/TO (Invitrogen, Carlsbad, CA, USA). Adenovirus (Ad)-mouse NGN3-enhanced green fluorescence protein (EGFP) and Ad-hamster NEUROD1-EGFP have been reported previously [21]. The pCB6-skin 16 mammalian expression vector bearing the mouse Ovol1 coding sequence has been described previously [15].
Animals
Partial pancreatectomy was performed as described previously [22] in young adult Sprague–Dawley rats (90%) and C57BL/6 mice (70–80%) (Taconic Laboratories, Germantown, NY, USA). After 3 or 4 days animals were killed under anaesthesia for sectioning of pancreases for immunohistochemistry. For embryonic pancreases, noon of the day the vaginal plug was detected was designated E0.5. All animal procedures were approved by the Joslin Institutional Animal Care and Use Committee.
Chromatin immunoprecipitation cloning
AR42J-B13 cells were plated in 100 mm cell culture dishes and infected for 4 h with AdNGN3-EGFP, AdNEUROD1-EGFP or Ad-EGFP at a multiplicity of infection of 20; medium was replaced and cells were maintained for 48 h. ChIP-cloning experiments were performed with minor modification to the suggested protocol (EZ-ChIP Kit; Upstate, Temecula, CA, USA). To increase the percentage of clones containing AR42J-B13 fragments, we used protein-G magnetic beads without adding blocking DNA to purify protein-DNA complexes. Experiments were performed in duplicate; the ChIP-cloning protocol was repeated twice. Plasmids from ∼50 to 80 colonies from AdNGN3-EGFP-infected cells and ∼20 to 30 colonies from Ad-EGFP-infected cells were isolated and sequenced. The sequence of each insert was compared against the rat genome database using BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) to determine the identity/location of each insert. Sequences were then analysed for the presence of DNA binding motif for the basic helix–loop–helix (bHLH) family of transcription factors (E-box, CANNTG).
Cloning and mutagenesis of the rat 1.1 kb Ovol1 promoter
The 1.1 kb sequence encompassing the 5′ region from −841 to +230 of the rat Ovol1 gene containing the E-box 1 was amplified by PCR using rat genomic DNA as template and cloned into the pGL2-Basic vector. A point mutation in the E-box 1 was created using a mismatch in the mutagenic primer and a kit (Phusion Site-Directed Mutagenesis Kit; New England Biolabs, Ipswich, MA, USA).
PCR analysis
Primers for quantitative PCR and RT-PCR experiments to detect pulled-down DNA fragments from ChIP experiments are in Electronic supplementary material (ESM) Table 1. The effect of bHLH factors on the expression of Ovol1 was determined using real-time PCR and SYBR Green Universal PCR Mix (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s protocol with tubulin as control; quantification used the ΔΔCt method [23].
Transient transfection analysis
AR42J-B13 or INS-1 cells (5 × 105) were plated into six-well plates the day before transfection and then transfected using 2.5 μl lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) added to 2 μg total mixture of vectors (containing 1 μg pGL2-OVOL1Prom1 [wild-type] or pGL2-OVOL1Prom1Mut [mutant], and 100 ng pcDNA5/TO-NGN3 or pcDNA5/TO-NEUROD1 [mammalian expression vectors] or pCMV-MYOD, with or without 100 ng pcDNA-E47 and 100 ng pRL-TK; pcDNA5/TO was used to balance the amount of DNA). Cells were incubated for 24 h at 37°C, lysed and luciferase activity measured using a kit (Dual Luciferase Kit; Promega, Madison, WI, USA) and Monolight 3010 Luminometer (BD Biosciences, San Jose, CA, USA).
Western blot analysis
Lysates from AR42J-B13 cells, acinar tissue, INS-1 cells, islets and testis (10–20 μg protein) were separated on 15% SDS-acrylamide/bis-acrylamide gels and transferred on to nitrocellulose membranes. Lysates from 293T cells that were either not transfected or transfected with Ovol1 expression plasmid were prepared and used as an additional control. OVOL1 and β-actin (rabbit anti-OVOL1, 1:1,000; Abcam, Cambridge, MA, USA; goat anti β-actin, 1:5,000; Santa Cruz Biotechnology, Santa Cruz, CA, USA) were detected using a chemiluminescence reagent (Western Lightning; Applied Biosystems, Foster City, CA, USA).
Immunohistochemistry
Paraffin sections of pancreases fixed in 4% (wt/vol.) paraformaldehyde were immunostained after microwaving in citrate acid buffer for antigen retrieval using primary antibodies shown in ESM Table 2. Secondary antibodies were from Jackson Immunolabs (West Grove, PA, USA). DAPI was used for nuclear staining. Peroxidase (Elite ABC kit; Vector Labs, Burlingame, CA, USA) with 3,3′-diaminobenzidine (Sigma, St Louis, MO, USA), a tyramide signal amplification fluorescein system (Perkin Elmer, Waltham, MA, USA) and biotin-streptavidin conjugation methods were used. Sections were examined on a BH-2 microscope (Olympus, Center Valley, PA, USA) or in confocal mode on an LSM 410 microscope (Zeiss, Tornwood, NY, USA). To compare OVOL1 intensity in embryonic pancreases (ESM Fig. 1), all sections were stained and photographed in parallel with identical settings. Final images were compiled using Adobe Photoshop.
Statistical analysis
For luciferase reporter and quantitative PCR assays, data are presented as mean+SD with statistic comparison of individual experimental treatments conducted using two-tailed Student’s t test. A p value <0.05 was considered significant.
Results
To identify direct targets of NGN3, we used ChIP-cloning to isolate protein-bound DNA fragments precipitated by anti-NGN3 antibody from the pancreatic cell line AR42J-B13 infected with NGN3-expressing Ad; these fragments were cloned. ChIP-cloning using anti-NGN3 antibody generated 50 to 80 colonies from cells infected with AdNGN3-EGFP and 20 to 30 colonies from those with control Ad-EGFP. The amount of DNA pulled down by control IgG was negligible and generated only a few colonies each. Hence we have only described analysis of clones identified using anti-NGN3 antibody.
Sequence analysis of insert fragments demonstrated that half of the 80 colonies from AdNGN3-EGFP-treated cells had at least one E-box. The cloned E-box-containing fragments ranged from 1,000 to 1,200 bp. Five independent clones contained the sequence of bHLH neurogenic factor 4 promoter region, five contained the sequence of the zinc-finger transcription factor Ovol1 and four contained the sequence of sex-determining region Y box 9 (Sox9) promoters. In contrast, sequence analysis of clones from AdEGFP-treated cells showed only one of 20 clones contained an E-box and none were transcription factors. Analysis of the 5′ flanking region of the rat Ovol1 gene with MatInspector software (www.genomatix.de/online_help/help_matinspector/matinspector_help.html) identified an E-box at −697 bp (E-box 1), present in the fragment cloned by ChIP-cloning approach, as well as three additional putative E-boxes at −1,177 bp (E-box 2), −1,209 bp (E-box 3) and −1,339 bp (E-box 4; Fig. 1a). Primers for E-Box1 region of Ovol1 promoter selectively amplified the PCR product with ChIP fragments from anti-NGN3-treated cells as template, confirming Ovol1 as a direct target of NGN3 (Fig. 1b).
NGN3 binds to Ovol1 promoter. a Schematic representation of rat Ovol1 gene showing location of four exons (E1–E4) and 5′ untranslated region. The portion of the promoter containing the four E-Boxes (E-Box 1 to 4), the DNA fragment cloned using ChIP cloning strategy (light grey vertical bars surrounding E-box 1) and the location of ChIP PCR primers are shown. Primer pair used for quantitative PCR (qPCR) amplifies a portion of exon 2 (E2); the primer pair for RT-PCR amplifies the PCR product encompassing regions of exons 3 and 4 (E3 and E4) that contain zinc fingers (upright black bars between E3 and E4). CDS, coding sequence. b In vivo binding of NGN3 to Ovol1 promoter fragment containing E-box 1. The cross-linked sonicated DNA–protein complex (input) or DNAs from immunoprecipitation using anti-NGN3 or IgG were used in PCR amplification with Ovol1-specific ChIP primers (see above [a] and ESM Table 1)
The role of OVOL1 in pancreas is not known. We therefore examined whether Ovol1 mRNA is expressed in pancreatic cells using total RNA prepared from AR42J-B13, INS-1 and rat islets. We also examined levels of OVOL1 protein in whole-cell extracts prepared from AR42J-B13, rat acinar tissue, INS-1 and rat islets; extracts of rat testis were used as positive control. Ovol1 mRNA (data now shown) and OVOL1 protein (Fig. 2a) were detected in all. The specificity of anti-OVOL1 antibody to recognise OVOL1 was confirmed by its ability to detect OVOL1 in previously reported tissue (testis) and in whole-cell extracts prepared from 293T cells transfected with a plasmid expressing Ovol1 coding sequence, but not in those from untransfected 293T cells. Anti-OVOL1 antibody detected protein bands of different intensities and different mobility in these extracts. Since, there are at least eight serines, threonines and tyrosines in the OVOL1 that can be phosphorylated by different protein kinases, we suggest that the different OVOL1 bands represent different post-translational modification of this protein in different cell types. By immunostaining, OVOL1 was observed throughout the ductal tree (including common pancreatic ducts, main ducts and small ductules), in islets and (patchily) in acini from adult rat (Fig. 2b) and mouse (Fig. 2c–e) pancreas. Although we isolated OVOL1 as a target of NGN3, which is observed only in endocrine progenitors, detection of OVOL1 in all major pancreatic cell types suggests that it may also have a role in adult pancreatic cell types.
OVOL1 is detected in multiple pancreatic-derived cell lines and adult rodent pancreas. a Immunoblot analysis using anti-OVOL1 antibody on whole-cell lysates from AR42J-B13, acinar tissue, INS-1, rat islets and testis as positive control. A protein band of ∼45 kDa corresponding to OVOL1 was detected in pancreatic cell lines, acinar, islets and testis. The specificity of OVOL1 antibody was verified by immunoblot for OVOL1 in 293T cells transfected with a plasmid expressing Ovol1 coding sequence, as well as untransfected 293T cells. b Within the adult rat pancreas OVOL1 protein (brown) showed strong signals in nuclei of pancreatic islets (I) and ducts (d). Its levels in acinar cells were patchy, with some cells showing no OVOL1 staining (black arrows) while several had strong levels (red arrows). c–e Similarly in adult mouse pancreas, OVOL1 (green) was present in almost all beta cells (c) (insulin in red), alpha cells (d) (glucagon in red), ducts (d) and in acini (e) (amylase in red). Scale bar, 50 μm
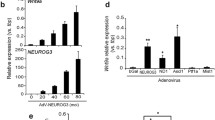
Basic HLH factors, such as NGN3, bind to the E-box as heterodimers with ubiquitously present bHLH proteins, such as E47, E12 and HEB [8, 24]. To confirm the ability of NGN3 to regulate Ovol1 expression, luciferase reporter constructs containing wild-type Ovol1 promoter sequence or its derivative with a mutated E-box 1 were co-transfected with Ngn3 and E47 expression vectors in AR42J-B13 cells. The presence of NGN3 and E47 together caused a fivefold reduction in luciferase activity from wild-type but not from the mutated E-box construct, demonstrating that NGN3 inhibits Ovol1 expression via the E-box 1 element (Fig. 3a). The effect of different bHLH factors was also examined to assess the specificity of inhibition of endogenous Ovol1 expression. Inducing the production of either NGN3 or NEUROD1 in AR42J-B13 and INS-1 cells reduced endogenous Ovol1 mRNA, but overexpression of muscle-specific bHLH factor MYOD did not (Fig. 3b). Reduced OVOL1 protein in extracts from cells transfected with NGN3 and NEUROD1 but not MYOD confirmed the selectivity of pancreatic bHLH factors to inhibit Ovol1 expression (data not shown).
NGN3 and NEUROD1 but not MYOD repress Ovol1 transcription via E-Box 1. a Luciferase activities were measured in AR42J-B13 cells 48 h after transfection with the indicated combination of reporter vectors (pGL2-OVOL1Prom1; wild-type or mutant), expression vectors (pcDNA-NGN3 and pcDNA-E47) and Renilla luciferase plasmid. The results are presented as relative to the luciferase activity of OVOL1 WT LUC (black bars) or mutant E1m LUC promoter (grey bars) in the absence of expression plasmids as 100%. The co-transfection of an Ngn3-expressing vector inhibited activity from the WT LUC reporter but not from the reporter containing mutation in the E-box 1 sequence in three independent experiments. b Total RNA from AR42J-B13 (black bars) and INS-1 (grey bars) cells transfected with the indicated combination of Ngn3, Neurod1 or Myod expression vectors with or without E47 for 48 h was analysed by quantitative PCR. Levels of Ovol1 mRNA in cells transfected with control vector (pcDNA/5TO) were used as 100%. Only pancreas-specific bHLH transcription factors (NGN3 and NEUROD1) significantly reduced the expression of endogenous Ovol1 mRNA. Values represent average ± SD of three independent experiments; *p < 0.05
As shown above, Ovol1 is a direct target of NGN3, which is expressed in endocrine progenitors in a narrow time frame [1, 2], suggesting that Ovol1 expression should increase as NGN3 level decreases. At E12.5, OVOL1 protein was undetectable in pancreatic and duodenal homeobox 1-positive pancreatic epithelium (Fig. 4a, b); at E15.5, when a significant number of cells with high levels of NGN3 were seen, only a few OVOL1-positive cells were observed (Fig. 4c). At E15.5, NGN3+ cells, which are mainly located in ‘trunk areas’ of ductal epithelium, showed almost no OVOL1 levels (Fig. 4c), but some cells in ‘tip areas’ with amylase+ differentiating acini (with no NGN3) showed OVOL1. At E18.5, when the number of NGN3-producing cells was reduced, the OVOL1 level was substantially higher in amylase+ acini (Fig. 4d) and insulin+ cells close to ducts (ESM Fig. 1). At this stage, NGN3+ cells were either low in OVOL1 or OVOL1-negative. Closer to birth, increased OVOL1 levels, both in intensity and number of positive cells, were seen (ESM Fig. 1). Overall, as NGN3 levels decreased in differentiating endocrine cells, OVOL1 levels increased. At E15.5 the number of cells with low OVOL1 was greater than can be accounted for by endocrine progenitors or endocrine cells; and with age, OVOL1 was seen in all pancreatic cell types. Thus, OVOL1 is present in differentiating precursors of all three pancreatic cell types.
OVOL1 is detected from secondary transition and negatively correlated with NGN3 in developing pancreas. a, b In adjacent sections from E12.5 mouse pancreas, PDX1-positive (a) (green) pancreatic epithelium, which is marked (a, b) by E-cadherin (red), showed no nuclear OVOL1 staining (green) (b) (green speckles staining artefact). Nuclei are labelled with DAPI in blue. Scale bar 100 μm. c, d In sections of embryonic pancreas stained for OVOL1 (green) and NGN3 proteins (red), more NGN3+ cells were detected at E15.5 (c) than at E18.5 (d). None of the NGN3-positive cells (white arrows) (c) co-expressed OVOL1 at E15.5. However, at E18.5 (d), two populations of NGN3+ cells, with either no (white arrow) or low (yellow arrows) OVOL1 were detected. Blue (c, d), amylase. Scale bars, 50 μm
In hair follicle and reproductive systems, OVOL1 inhibits proliferation of stem/progenitor cells and enhances their differentiation into terminally differentiated cell types [15]. Therefore, we examined whether the levels of OVOL1 changed in concert with the balance between pancreatic proliferation and differentiation. By E18.5, OVOL1 levels significantly increased in amylase+ tip cells, which are quiescent. Interestingly, there are several populations in epithelial cord (duct) cells and centro-acinar cells that may serve as progenitors. One of these was positive for Ki67 and not for OVOL1 (Fig. 5a), another was positive for Ki67 and weak for OVOL1 (Fig. 5a) and a third was strongly positive for OVOL1 and weak for Ki67 (Fig. 5a, ESM Fig. 2). This observation suggests that, in progenitor cells, OVOL1 may regulate a balance between proliferation and differentiation, while differentiated cells only show OVOL1. Consistent with the inverse association between Ki67 and OVOL1 in differentiated cells, we observed that a significant proportion of quiescent cells in adult pancreas expressed high levels of OVOL1 (Fig. 5b), while the occasional Ki67+ islet cells in adult animals were negative for OVOL1.
OVOL1 and Ki67 are inversely detected in embryonic, adult and regenerating pancreas of partial pancreatectomised rodent. a In E18.5 mouse pancreas, there were several populations in epithelial cord (duct) cells and centro-acinar cells, which may serve as progenitors: one positive for Ki67 (red) and negative for OVOL1 (white arrows) (green), another positive for Ki67 and weakly positive for OVOL1 (yellow arrows), and a third highly positive for OVOL1 and weakly positive for Ki67 (blue arrows). For split channel images of this panel, see ESM Fig. 2. b In adult mouse islets, OVOL1 (green) was detected in almost all insulin (blue)-positive cells. However, a Ki67+ cell (red, yellow arrow) was negative for OVOL1. Inset: a high-resolution split channel image of this single Ki67+ OVO1− insulin+ cell. Four days after pancreatectomy (c) rat and (d) mouse, OVOL1-positive (green) cells were seen in acini in remnant, but rarely detected in proliferating ducts (indicated by * [c] and white arrows [d]; Ki67+ ductules, red) in regenerating foci, indicating that OVOL1 levels increase with differentiation. Blue (d), amylase. Scale bar, 50 μm
To further explore this inverse correlation of OVOL1 with proliferation, we examined OVOL1 levels after partial pancreatectomy, a well-characterised model of pancreatic regeneration [22]. At 3 to 4 days post surgery, newly forming pancreatic lobes (foci of regeneration or focal areas) are seen with proliferating ductules (young areas enriched in ducts) and newly differentiating cells [25]. The early stages of these new lobes are seen as small proliferating ductules that have many markers of pancreatic progenitors [26]. OVOL1 levels were high in remnant pancreas, but very low in ductal structures in these focal areas (Fig. 5c, d). These data are consistent with high OVOL1 levels in quiescent adult cells and low levels in proliferating cells (Fig. 5a). Thus, similar to its role in other tissues, Ovol1 expression in pancreas is inversely associated with proliferation.
Discussion
NGN3 is a key transcription factor of endocrine differentiation, so identification and characterisation of its targets would be an important step towards regulating differentiation of beta cells. Using a ChIP-cloning approach to search for direct targets of NGN3, we identified a zinc-finger transcription factor OVOL1. Ovo is an evolutionarily conserved family of genes that is found in C. elegans, Drosophila, mice and humans, and regulates development of epithelial tissues and germ cells [27–29]. In mice, Ovol1 has a limited cellular distribution, with expression reported in skin, testis and kidneys [15, 27, 30]. OVOL1 often acts as a transcriptional repressor that promotes transition of cells from a proliferating, less differentiated state to a post-mitotic, more differentiated state [15]. This property of OVOL1 is appropriate for it being a downstream target of NGN3, a transcription factor that regulates conversion of pancreatic progenitors (proliferating, less differentiated) into endocrine cells (post-mitotic, more differentiated). During development, transient production [2, 4] of NGN3 is sufficient to induce production of several transcription factors that then regulate endocrine differentiation [6, 8]. While NGN3 activates most of its characterised targets, it can inhibit transcription both of itself [31] and of Ovol1. Additionally, bHLH factor NEUROD1, but not MYOD, also inhibits Ovol1 expression via E-box 1 (Fig. 3 and data not shown), suggesting that other pancreas-enriched bHLH factors expressed in ductal and acinar cells may regulate Ovol1 expression in these cell types. NEUROD1 is likely to regulate postnatal expression of Ovol1 in endocrine cells, while NGN3 may play the primary role during embryonic development and neogenesis after birth [32].
Recent large-scale analyses of gene expression have successfully identified several downstream targets of NGN3 [12–14]. In these studies, the detection of NGN3 targets was based on the identification of genes expressed in NGN3+ but not in NGN3− pancreatic cells during embryonic development, a process that is unlikely to identify genes inhibited by NGN3, such as Ovol1. Consistent with this prediction, Ovol1 was not identified as a differentially expressed gene in these studies [8, 12]; this lack could in part be due to the low expression of Ovol1 and/or absence of an Ovol1 probe set in the arrays used, as well as to expression of Ovol1 in all pancreatic cell types. Petri and colleagues [14] compared wild-type and NGN3 knockout pancreas from E13.5 and E15.5 using ArrayTAG 20 K murine gene collection spotted oligos. They identified only a few genes that were upregulated in Ngn3 knockout mice at E15.5, a time when only occasional cells express Ovol1. Another study [13] that used a similar experimental strategy analysed gene expression data at E18.5. Re-analysis of these results showed a twofold increase in Ovol1 expression at E18.5 when NGN3 levels are reduced (data not shown), which is consistent with our conclusion that Ovol1 is a downstream target of NGN3.
Insights on the function of OVOL1 have been gained from Ovol1 null mice. On a mixed genetic background, Ovol1-deficient mice are small, produce aberrant hairs, have abnormalities in kidney and display hypogenitalism with a reduced ability to reproduce [27]. The developing epidermis of Ovol1 −/− mice fails to properly restrict the proliferative potential of progenitor cells and cultured keratinocytes failed to undergo growth arrest in response to extrinsic growth-inhibitory signals. Possible mechanisms involved include upregulation of c-myc expression, which is directly inhibited by OVOL1 in Ovol1-deficient suprabasal cells [15]. In the testis, Ovol1-deficient germ cells are defective in progressing through the pachytene stage and have upregulated expression of Id2, a known regulator of spermatogenesis that is directly inhibited by OVOL1 [30]. These OVOL1 targets, inhibitor of differentiation 2 (ID2) and c-Myc, have known effects in the pancreas. Thus ID2 protein inhibits NEUROD1 binding to its target E-box [33] and c-Myc represses insulin transcription while promoting proliferation of beta cells [34], thereby controlling the inverse relationship between proliferation and differentiation in pancreatic endocrine cells. By regulating levels of transcription factors such as c-Myc and ID2, OVOL1 could inhibit proliferation of progenitors and enhance their differentiation into mature cell types. Preliminary analysis of pancreases from litters containing 3-week-old Ovol1 wild-type, heterozygous and null animals on C57/BL6J background showed increased Ki67-positive cells in all pancreatic compartments in the Ovol1-deficient compared with control pancreas (data not shown). One of the limitations of this analysis was that on a pure C57/BL6J background, survival of Ovol1 knockouts was reduced both from E15.5 to E18.5 and during the first two postnatal weeks [35]. Hence we cannot discriminate between the loss of Ovol1 function vs any survival effect. A systematic analysis of Ovol1 knockout mice will be required to confirm the role of this factor in the pancreas.
In summary, we report the identification of a novel NGN3 target, Ovol1, which represents a new class of genes inhibited by NGN3. We show that during embryonic development Ovol1 expression is inversely correlated with that of NGN3 and Ki67. A similar inverse correlation between OVOL1 and Ki67 levels exists during pancreatic regeneration. We show that Ovol1 is broadly expressed in all pancreatic cell types in late embryos and adults, and that its expression is regulated by pancreatic bHLH factors. Our findings that Ovol1 expression is initiated during secondary transition and is enhanced in differentiated but not in dividing cells suggest that OVOL1 may regulate the balance between proliferation and differentiation of pancreatic progenitors. In adults, OVOL1 may play an important role in maintaining the quiescence/differentiated state of pancreatic cells. Further characterisation of this factor could enable it to be used to enhance the proliferative potential of embryonic and adult pancreatic progenitors.
Abbreviations
- Ad:
-
Adenovirus
- bHLH:
-
Basic helix–loop–helix
- ChIP:
-
Chromatin immunoprecipitation
- E:
-
Embryonic day
- EGFP:
-
Enhanced green fluorescence protein
- ID2:
-
Inhibitor of differentiation 2
- NEUROD1:
-
Neurogenic differentiation 1
- NGN3:
-
Neurogenin-3
- NKX2.2:
-
NK2 transcription factor related, locus 2
- OVOL1:
-
OVO homologue-like 1
- PAX4:
-
Paired box gene 4
References
Apelqvist A, Li H, Sommer L et al (1999) Notch signalling controls pancreatic cell differentiation. Nature 400:877–881
Gradwohl G, Dierich A, LeMeur M, Guillemot F (2000) Neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc Natl Acad Sci U S A 97:1607–1611
Grapin-Botton A, Majithia AR, Melton DA (2001) Key events of pancreas formation are triggered in gut endoderm by ectopic expression of pancreatic regulatory genes. Genes Dev 15:444–454
Schwitzgebel VM, Scheel DW, Conners JR et al (2000) Expression of neurogenin3 reveals an islet cell precursor population in the pancreas. Development 127:3533–3542
White P, May CL, Lamounier RN, Brestelli JE, Kaestner KH (2008) Defining pancreatic endocrine precursors and their descendants. Diabetes 57:654–668
Smith SB, Gasa R, Watada H, Wang J, Griffen SC, German MS (2003) Neurogenin3 and hepatic nuclear factor 1 cooperate in activating pancreatic expression of Pax4. J Biol Chem 278:38254–38259
Watada H, Scheel DW, Leung J, German MS (2003) Distinct gene expression programs function in progenitor and mature islet cells. J Biol Chem 278:17130–17140
Huang HP, Liu M, El-Hodiri HM, Chu K, Jamrich M, Tsai MJ (2000) Regulation of the pancreatic islet-specific gene BETA2 (neuroD) by neurogenin 3. Mol Cell Biol 20:3292–3307
Haumaitre C, Lenoir O, Scharfmann R (2008) Histone deacetylase inhibitors modify pancreatic cell fate determination and amplify endocrine progenitors. Mol Cell Biol 28:6373–6383
Gu G, Dubauskaite J, Melton DA (2002) Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development 129:2447–2457
Sugiyama T, Rodriguez RT, McLean GW, Kim SK (2007) Conserved markers of fetal pancreatic epithelium permit prospective isolation of islet progenitor cells by FACS. Proc Natl Acad Sci U S A 104:175–180
Gu G, Wells JM, Dombkowski D, Preffer F, Aronow B, Melton DA (2004) Global expression analysis of gene regulatory pathways during endocrine pancreatic development. Development 131:165–179
Juhl K, Sarkar SA, Wong R, Jensen J, Hutton JC (2008) The mouse pancreatic endocrine cell transcriptome defined in the embryonic Ngn3 null mouse. Diabetes 57:2755–2761
Petri A, Ahnfelt-Ronne J, Frederiksen KS et al (2006) The effect of neurogenin3 deficiency on pancreatic gene expression in embryonic mice. J Mol Endocrinol 37:301–316
Nair M, Teng A, Bilanchone V, Agrawal A, Li B, Dai X (2006) Ovol1 regulates the growth arrest of embryonic epidermal progenitor cells and represses c-myc transcription. J Cell Biol 173:253–264
Mellitzer G, Bonne S, Luco RF et al (2006) IA1 is NGN3-dependent and essential for differentiation of the endocrine pancreas. EMBO J 25:1344–1352
Graham FL, Smiley J, Russell WC, Nairn R (1977) Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol 36:59–74
Kieffer TJ, Heller RS, Unson CG, Weir GC, Habener JF (1996) Distribution of glucagon receptors on hormone-specific endocrine cells of rat pancreatic islets. Endocrinology 137:5119–5125
Mashima H, Shibata H, Mine T, Kojima I (1996) Formation of insulin-producing cells from pancreatic acinar AR42J cells by hepatocyte growth factor. Endocrinology 137:3969–3976
Sharma A, Moore M, Marcora E et al (1999) The NeuroD1/BETA2 sequences essential for insulin gene transcription colocalize with those necessary for neurogenesis and p300/CREB binding protein binding. Mol Cell Biol 19:704–713
Yatoh S, Akashi T, Chan PP et al (2007) NeuroD and reaggregation induce beta-cell specific gene expression in cultured hepatocytes. Diabetes Metab Res Rev 23:239–249
Bonner-Weir S, Trent DF, Weir GC (1983) Partial pancreatectomy in the rat and subsequent defect in glucose-induced insulin release. J Clin Invest 71:1544–1553
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408
Sharma A, Henderson E, Gamer L, Zhuang Y, Stein R (1997) Analysis of the role of E2A-encoded proteins in insulin gene transcription. Mol Endocrinol 11:1608–1617
Bonner-Weir S, Baxter LA, Schuppin GT, Smith FE (1993) A second pathway for regeneration of adult exocrine and endocrine pancreas. A possible recapitulation of embryonic development. Diabetes 42:1715–1720
Sharma A, Zangen DH, Reitz P et al (1999) The homeodomain protein IDX-1 increases after an early burst of proliferation during pancreatic regeneration. Diabetes 48:507–513
Dai X, Schonbaum C, Degenstein L, Bai W, Mahowald A, Fuchs E (1998) The ovo gene required for cuticle formation and oogenesis in flies is involved in hair formation and spermatogenesis in mice. Genes Dev 12:3452–3463
Johnson AD, Fitzsimmons D, Hagman J, Chamberlin HM (2001) EGL-38 Pax regulates the ovo-related gene lin-48 during Caenorhabditis elegans organ development. Development 128:2857–2865
Oliver B, Perrimon N, Mahowald AP (1987) The ovo locus is required for sex-specific germ line maintenance in Drosophila. Genes Dev 1:913–923
Li B, Nair M, Mackay DR et al (2005) Ovol1 regulates meiotic pachytene progression during spermatogenesis by repressing Id2 expression. Development 132:1463–1473
Lee JC, Smith SB, Watada H et al (2001) Regulation of the pancreatic pro-endocrine gene neurogenin3. Diabetes 50:928–936
Xu X, D'Hoker J, Stange G et al (2008) Beta cells can be generated from endogenous progenitors in injured adult mouse pancreas. Cell 132:197–207
Ghil SH, Jeon YJ, Suh-Kim H (2002) Inhibition of BETA2/NeuroD by Id2. Exp Mol Med 34:367–373
Laybutt DR, Weir GC, Kaneto H et al (2002) Overexpression of c-Myc in beta-cells of transgenic mice causes proliferation and apoptosis, downregulation of insulin gene expression, and diabetes. Diabetes 51:1793–1804
Teng A, Nair M, Wells J, Segre JA, Dai X (2007) Strain-dependent perinatal lethality of Ovol1-deficient mice and identification of Ovol2 as a downstream target of Ovol1 in skin epidermis. Biochim Biophys Acta 1772:89–95
Acknowledgements
This study was supported by the NIH P30 DK36836 Joslin Diabetes and Endocrinology Research Center (DERC) Advanced Microscopy Core, JDRF Research Grant (to A. Sharma), the Diabetes Research and Wellness Foundation, and by a group of private donors. W.-C. Li was supported by a JDRF Post Doctoral Fellowship, W. Nishimura by the Mary K. Iacocca Fellowship and K. Juhl by a Lundbeckfonden fellowship. Mouse monoclonal anti-NGN3 antibody developed by O. D. Madsen was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by Department of Biological Sciences, University of Iowa, Iowa, IA, USA.
Duality of interest
The authors declare that there is no duality of interest associated with this manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
A. Vetere and W.-C. Li contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM Table 1
(PDF 14 kb)
ESM Table 2
(PDF 10 kb)
ESM Fig. 1
OVOL1 levels increase in developing pancreas towards birth. (A) Few OVOL1low cells (in green, white arrows) can be detected at E15.5. (B, C) From E15.5 until birth, OVOL1 levels increase and begins to be detectable in insulin+ (red) cells and adjacent ductal (d) structures. The images were stained and photographed in parallel at the same confocal settings to obtain a direct comparison of intensity of OVOL1 levels (PDF 748 kb)
ESM Fig. 2
Differential levels of Ki67 protein in OVOL1+ cells in E18.5 mouse pancreas. Figure 5a is shown here as split channel images, so that it is clear that the replicating cells (Ki67+) are in the trunk with some OVOL1 negative (white arrows) and others producing some OVOL1 protein (yellow arrows). A third population in the trunk are cells with high levels of OVOL1 protein with weak Ki67 (blue arrows). Strongly positive OVOL1+ cells are mainly detected in tip area where most acinar cells are quiescent (PDF 369 kb)
Rights and permissions
About this article
Cite this article
Vetere, A., Li, WC., Paroni, F. et al. OVO homologue-like 1 (Ovol1) transcription factor: a novel target of neurogenin-3 in rodent pancreas. Diabetologia 53, 115–122 (2010). https://doi.org/10.1007/s00125-009-1567-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-009-1567-5