Abstract
Aims/hypothesis
Mutations in the HNF1A (previously known as TCF1) gene encoding hepatocyte nuclear factor-1α (HNF1A) lead to the development of maturity-onset diabetes of the young, type 3 (HNF1A-MODY), characterised by impaired insulin secretion and a reduction in beta cell mass. HNF1A plays an important role in pancreatic beta cell differentiation and survival. The mammalian target of rapamycin (mTOR) is a central growth factor- and nutrient-activated protein kinase controlling cell metabolism, growth and survival. We investigated the role of mTOR inactivation in the decline in beta cell mass in a cellular model of HNF1A-MODY.
Methods
Previously we showed that suppression of HNF1A function via expression of a dominant-negative mutant (DN-HNF1A) decreases insulin gene transcription in insulinoma (INS-1) cells. We investigated the signalling of two distinct mTOR protein complexes, mTORC1 and mTORC2, in response to DN-HNF1A induction.
Results
We observed delayed inactivation of mTORC2 48 h after DN-HNF1A induction, evidenced by a reduction in serine 473 phosphorylation of thymoma viral proto-oncogene 1 (AKT1). We also observed an early inactivation of mTORC1 24 h after DN-HNF1A induction, which was detected by decreases in threonine 389 phosphorylation of p70 ribosomal protein S6 kinase (S6K1) and serine 65 phosphorylation of translational inhibitor eukaryotic translation initiation factor 4E binding protein 1 (4E-BP1). Flow cytometry and gene expression analysis demonstrated a pre-apoptotic decrease in INS-1 cell size in response to DN-HNF1A induction, and an increase in the level of the mTORC1-regulated cell-cycle inhibitor, cyclin-dependent kinase inhibitor 1B p27.
Conclusions/interpretation
Our data suggest that mTOR kinase and signalling through mTORC1 are highly sensitive to suppression of HNF1A function, and may contribute to disturbance of cell-size regulation and cell-cycle progression in HNF1A-MODY.
Similar content being viewed by others
Introduction
MODY is an autosomal dominant and monogenic form of diabetes characterised by early age of onset and primary pancreatic beta cell dysfunction [1]. Hepatocyte nuclear factor-1α (HNF1A) MODY is believed to be the commonest form of MODY [2] and results from heterozygous loss-of-function mutations of transcription factor HNF1A (previously known as TCF1) [3, 4]. HNF1A is produced in the liver, kidney, intestine and pancreatic islets [5]. It participates in a transcription factor network that regulates pancreatic development as well as fatty acid, protein and carbohydrate metabolism [6]. This network has also been shown to be critically involved in the pathophysiology of type 2 diabetes [7].
Beta cell transgenic mice carrying a deletion of the Hnf1a gene have defective glucose-stimulated insulin secretion without insulin resistance of target tissues [8], similar to individuals with HNF1A-MODY [9]. Animal models of HNF1A-MODY suggest a decline in functional beta cell mass as the key underlying mechanism of this defect [10]. A decrease in beta cell mass can be caused by defects in beta cell differentiation and proliferation, a decrease in beta cell size or increased beta cell apoptosis. Previous studies from our and other laboratories have shown that knockdown of Hnf1a or expression of dominant-negative mutants of Hnf1a (DN-HNF1A) decreases insulin release and insulin gene transcription, and inhibits the activation of the phosphoinositide 3-kinase (PI3K)/AKT1 pathway, thereby arresting cell proliferation and sensitising beta cells to apoptosis [11–13].
Reduced insulin signalling can also impact on mammalian target of rapamycin (mTOR) signalling. Originally identified in Saccharomyces cerevisiae [14], mTORs are PI3K-related Ser/Thr kinases. mTOR integrates signals from a variety of upstream sources, including nutrients such as amino acids and mitogenic and growth factors [15]. The signalling pathway initiated by growth factors involves insulin/IGF receptor-induced PI3K and AKT1 activation leading to phosphorylation of tuberous sclerosis 1 protein (TSC1)/tuberous sclerosis 2 protein (TSC2) and inhibition of RAS-homologue enriched in brain (RHEB)-GTPase [16]. mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2) are two structurally and functionally distinct protein complexes that share mTOR as their catalytic subunit. mTORC1 contains mTOR, regulator-associated protein of mTOR (Raptor), proline-rich Akt substrate of 40 kDa (PRAS40) and G protein β subunit-like protein (GBL) and regulates temporal aspects of cell growth, such as ribosomal biogenesis and protein synthesis. Activated mTORC1 directly phosphorylates ribosomal p70 S6 kinase (S6K1), leading to its activation and phosphorylation of the repressors of mRNA translation 4E-binding proteins (4E-BP), thereby causing their inactivation [17]. mTORC1 initiates a negative feedback loop to modify the activity of AKT1 through S6K1. mTORC1-activated kinase S6K1 phosphorylates IRS1 and IRS2 proteins, resulting in decreased IRS-protein stability in insulin/insulin-like growth factor-responsive cells [17]. Another mTOR complex, mTORC2, contains mTOR, rapamycin-insensitive companion of mTOR (Rictor), PRR5/proline-rich protein 5 (Protor), mammalian stress-activated protein kinase-interacting protein 1 (SIN1) and GBL, and appears to regulate long-term changes in cellular physiology such as spatial organisation [18]. mTORC2 also phosphorylates and activates AKT1 at the stimulatory Ser473 residue. In conjunction with phosphoinositide-dependent kinase-1 (PDK1)-mediated phosphorylation of Thr308, this modification drives the full activation of AKT1 [19, 20]. The TSC1–TSC2 complex was found to physically associate with mTORC2 and may regulate the mTORC2 phosphorylation of AKT1 [21].
Given the impact of dominant-negative suppression of HNF1A function on insulin signalling and the central role of mTOR in mediating growth-factor signalling, we investigated the potential role of mTOR in the decline in beta cell mass in HNF1A-MODY, using the in vitro model of INS-1 cells inducibly expressing DN-HNF1A.
Methods
Cultivation and treatment of insulinoma INS-1 cells overexpressing Hnf1a in an inducible system
Rat INS-1 insulinoma cells overexpressing wild-type Hnf1a (WT-HNF1A) or a dominant-negative mutant of Hnf1a (DN-HNF1A) (SM6) under control of a doxycycline-dependent transcriptional activator have been described previously [13, 22]. The SM6 mutant contains a substitution of 83 amino acids in the Hnf1a DNA-binding domain, resulting in the formation of non-functional heterodimers with endogenous HNF1A [23] (see Electronic supplementary material [ESM]).
Western blotting
Cells were rinsed with ice-cold PBS and lysed in buffer containing 62.5 mmol/l Tris–HCl (pH 6.8), 2% SDS (wt/vol.), 10% glycerin (wt/vol.) and protease inhibitor cocktail. Protein content was determined using the Pierce BCA Micro Protein Assay kit (Rockford, IL, USA). Samples were supplemented with 2-mercaptoethanol and denatured at 95°C for 5 min. An equal amount of protein (20–50 μg) was separated with 5–15% SDS-PAGE (wt/vol.) and blotted to nitrocellulose membranes (Protean BA 85; Schleicher & Schuell, Dassel, Germany). The blots were blocked with 5% non-fat milk (wt/vol.) in blocking solution (15 mmol/l Tris–HCl [pH 7.5], 200 mmol/l NaCl and 0.1% Tween-20 [wt/vol.]) for 2 h at room temperature. Membranes were incubated overnight at 4°C with the primary antibodies (see ESM).
Flow cytometry
After DN-HNF1A induction, INS-1 cells were collected with trypsin-EDTA and incubated in binding buffer (10 mmol/l HEPES, 135 mmol/l NaCl, 5 mm/l CaCl2) containing Annexin-V FITC (5 μl/ml) (BioVision, Mountain View, CA, USA) and propidium iodide (1 μg/ml) for 15 min at 37°C. Cells (1 × 105) were resuspended in ice-cold binding buffer and analysed on a Cyflow ML 16 flow cytometer (Partec, Münster, Germany) equipped with a 488, 405 and 532 nm laser. Annexin-V was excited with the 488 nm laser and fluorescence emission was collected in the FL1 channel through a 520 nm band pass filter; propidium iodide was excited with the 488 nm laser and fluorescence emission was collected in the FL3 channel through a 620 nm long pass filter. Gated cells (1 × 104) were acquired for each sample and analysed by using the Flowmax software (Partec, Münster, Germany). Cell-size analysis was performed on healthy cells gated on a forward scatter channel/FL1 scatter plot; apoptotic Annexin-V-positive and necrotic propidium iodide-positive cells were therefore excluded from the analysis, and the median values of the forward scatter of the remaining populations used a measure of cell size.
Real-time quantitative PCR
Expression of Raptor Rictor, Sin1 (also known as Mapkap1), mTor (also known as Frap1), cyclin-dependent kinase inhibitor 1B (p27 [also known as Cdkn1b]) and L-type pyruvate kinase (Pklr) mRNA was examined using real-time quantitative PCR (qPCR). INS-1 cells were harvested at the appropriate time-points and total RNA was extracted using the RNeasy mini Kit (Qiagen, Crawley, UK). First-strand cDNA synthesis was performed with 2 μg total RNA as template using Superscript II reverse transcriptase (Invitrogen, Paisley, UK) primed with 50 pmol random hexamers (New England Biolabs, Ipswich, MA, USA). qPCR was performed using the LightCycler 2.0 (Roche Diagnostics, Indianapolis, IN, USA) and the QuantiTech SYBR Green PCR kit (Qiagen) as per manufacturers’ protocols and 25 pmol of primer pair concentration (Sigma-Genosys, Zwÿndrecht, the Netherlands). The PCR products were designed to be 150–200 bp in length. Specific primers for each gene analysed were designed using Primer3 software (http://frodo.wi.mit.edu/cgi-bin/ primer3/primer3_http://www.cgi; see ESM) [24]. Each primer pair was tested with a logarithmic dilution of a cDNA mix to generate a linear standard curve which was used to calculate the PCR efficiency and their specificity was determined by melting-curve analysis and gel electrophoresis of products. The PCR reactions were performed in a 20 μl volume. The PCR was performed as follows: there was an activation step of HotStartTaq polymerase at 95°C for 15 min, followed by 50 cycles of denaturation at 95°C for 15 s, annealing at 59°C for 20 s, and extension at 72°C for 20 s. The data were analysed using Lightcycler Software 4.0 with all samples normalised to β-actin.
Statistics
Data are given as means ± SE. For statistical comparison, ANOVA and subsequent Tukey’s test were employed. Results were considered to be statistically significant where p < 0.05.
Results
INS-1 cells inducibly expressing DN-HNF1A but not WT-HNF1A show an early decrease in Thr389 S6K1 phosphorylation
Expression of DN-HNF1A was induced in INS-1 cells by treatment with doxycycline for 12, 24, 36 and 48 h. Treatment with 500 ng/ml doxycycline led to a rapid induction of DN-HNF1A in INS-1 cells stably transfected with the respective doxycycline-dependent transcriptional activator (Fig. 1a). To explore alterations in mTOR signalling, the phosphorylation status of AKT1 (Ser473) and S6K1 (Thr389) was analysed. In agreement with our previous study [12], we found that induction of DN-HNF1A reduced mTORC2-mediated Ser473 phosphorylation of AKT1 after 48 h (see also Fig. 4c). This reduction in AKT1 Ser473 phosphorylation correlated with an increase in caspase-3 activity and apoptotic cell death in the cultures [12]. In contrast, mTORC1-dependent phosphorylation of S6K1 within the linker domain (Thr389) was significantly reduced as early as 24 h after induction of DN-HNF1A (Fig. 1b). This decrease persisted during the 48 h of DN-HNF1A induction. Dephosphorylation of S6K1 at Thr389 could not be detected in cells induced to produce WT-HNF1A (Fig. 1c). These results suggested a differential inactivation of mTORC1- and mTORC2-dependent signalling in response to DN-HNF1A expression.
DN-HNF1A induces an early decrease in mTORC1 signalling. a Time course of induction of DN-HNF1A in response to 500 ng/ml doxycycline. Immunodetection was performed using a HNF1A antibody. INS-1 cells were induced to overproduce DN-HNF1A (b) or WT-HNF1A (c) for 12, 24, 36 and 48 h. The activation status of p70 S6 kinase was detected by western blotting using a Thr389-phosphorylation-specific antibody, designated by P-Thr389. The nitrocellulose membrane was stripped and re-probed for total S6K1. Experiments were repeated twice with similar results
Early inactivation of mTORC1- and S6K1-dependent signalling pathways in response to DN-HNF1A
To further explore this finding we analysed the activation status of other mTORC1- and S6K1-dependent signalling pathways in response to DN-HNF1A expression. mTORC1 controls protein synthesis via phosphorylation of 4E-BP1 [25]. 4E-BP1 normally binds to and inhibits eukaryotic initiation factor 4 (eIF4E), thereby blocking cap-dependent translation initiation. INS-1 cells were induced to express DN-HNF1A over 48 h. Similar to the effects we observed regarding the activation status of S6K1, we observed a decrease in Ser65 phosphorylation of 4E-BP1 as early as 24 h after the induction of DN-HNF1A (Fig. 2a).
Early decrease in mTORC1-mediated 4E-BP1 phosphorylation and S6K1-mediated mTOR phosphorylation. INS-1 cells were induced to overproduce DN-HNF1A for 0, 12, 24, 36 and 48 h. a The activation status of 4E-BP1 was detected by western blotting using a Ser65-phosphorylation-specific antibody (P-Ser65). Membrane was stripped and re-probed with an anti-α tubulin antibody to prove equal loading of samples. b The activation of mTOR was detected by western blotting using a Ser2448 phosphorylation antibody (P-Ser2448). The experiment was repeated four times with similar results
We also investigated S6K1-dependent phosphorylation of mTOR. S6K1 is able to phosphorylate mTOR at Ser2448, a mechanism described as a feed-forward activation loop [26]. Using a phosphorylation-specific antibody we detected a reduction in Ser2448 mTOR phosphorylation 24 h after DN-HNF1A induction that correlated with the decrease in phosphorylation (Fig. 2b). Total protein levels of mTOR remained stable during the induction of DN-HNF1A. In contrast to the effects observed upon DN-HNF1A induction, WT-HNF1A INS-1 cells failed to exhibit any decrease in mTOR Ser2448 phosphorylation (data not shown).
Differential gene expression of mTorc1 and mTorc2 constituent’s Raptor, Rictor and Sin1 does not occur during the induction of DN-HNF1A
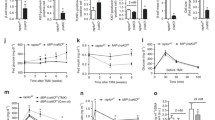
The differential effect of DN-HNF1A on mTORC1- and mTORC2-dependent signal transduction could be secondary to changes in gene expression of individual mTorc1 and mTorc2 constituents. While mTOR and Gbl are components of both mTorc1 and mTorc, changes in Raptor, Rictor and Sin1 gene expression upon induction of DN-HNF1A could manifest in differential mTORC activities. To address this question, cells were induced to express Hnf1a and RNA was isolated between 8 and 48 h following induction. Gene expression was analysed by qPCR relative to the expression of β-actin mRNA, which did not change over 48 h (data not shown). Control experiments detected a prominent reduction in mRNA levels of Pklr, a known Hnf1a target gene, in response to DN-HNF1A induction (Fig. 3a). Although we detected increased levels over time, we failed to detect any significant difference in expression levels of Raptor (constituent of mTorc1) compared with Rictor or Sin1 mRNA (constituents of mTorc2), suggesting that alterations in gene expression did not cause the differential inactivation kinetics of mTORC1 and mTORC2 (Fig. 3b–d). Levels of mTor mRNA remained largely unaffected by DN-HNF1A induction (Fig. 3e).
Quantification of gene expression of mTorc components during DN-HNF1A induction in INS-1 cells. INS-1 cells were treated with 500 ng/ml doxycycline from 0 to 48 h. Following treatment mRNA expression of a Pklr, b Raptor, c Rictor, d Sin1 and e mTOR was examined using real-time qPCR relative to β-actin. Expression levels were normalised to control cells and data shown are the means of n = 3 separate treatments. *p < 0.05 for difference from non-induced controls. Experiments were repeated three times with similar results
Effect of extracellular glucose on mTORC1-dependent S6K1 and mTORC2-dependent AKT1 phosphorylation
mTORC1 signalling is dependent on nutrient and energy supply, whereas comparatively little is known about the upstream regulation of mTORC2 [18, 21]. To explore the effect of nutrient supply on the differential inactivation of mTORC1- and mTORC2-dependent signalling in response to DN-HNF1A, we investigated the effect of extracellular glucose levels on the Thr389 phosphorylation status of S6K1 and the Ser473 phosphorylation status of AKT1. INS-1 cells were induced to express DN-HNF-1A for 24 or 48 h at a normal glucose concentration of 6 mmol/l, and were subsequently challenged with increasing glucose concentrations for 30 min. In agreement with previous reports, non-induced controls showed a glucose-dependent Thr389 phosphorylation of S6K1 (Fig. 4a). After induction of DN-HNF1A, the overall Thr389 phosphorylation of S6K1 was significantly reduced at all glucose concentrations tested (Fig. 4a). However, INS-1 cells expressing DN-HNF1A were still glucose responsive, as a 30 min treatment in 18 mmol/l extracellular glucose elevated the levels of Thr389 phosphorylated S6K1 to levels detectable in non-induced controls maintained at 6 mmol/l glucose. In contrast, mTORC2-dependent AKT-473 phosphorylation in non-induced and induced DN-HNF1A- or WT-HNF1A-expressing INS-1 cells was not affected by addition of increasing concentrations of extracellular glucose (Fig. 4b–d).
Effect of extracellular glucose on mTORC1-dependent S6K and mTORC2-dependent AKT1 phosphorylation. INS-1 cells were induced to overproduce DN-HNF1A for 24 h (a, b) or 48 h (c), and WT-HNF1A for 24 h (d) in the presence of 6 mmol/l glucose, and were then exposed to the indicated glucose concentrations for 30 min. The activation status of S6K1 or AKT1 was detected by western blotting using a Thr389- and Ser473-phosphorylation-specific antibody, respectively (P-Thr389, P-Ser473). Experiments were repeated twice with similar results
Induction of DN-HNF1A is associated with a reduction in INS-1 cell size
S6K1 phosphorylates the 40S ribosomal protein S6 and other proteins involved in the regulation of protein synthesis such as eukaryotic translation initiation factor 4B (eIF4B), eukaryotic elongation factor-2 kinase (eEF2K) and S6K1 specific interactor SKAR (S6K1 Aly/REF-like substrate) [27]. Via these activities, S6K1 has been shown to be of central importance in the control of cell size [18, 27]. We finally investigated whether the decrease in mTORC1 and S6K1 signalling also precipitated a reduction in INS-1 cell size during the induction of DN-HNF1A. Cells were induced to express DN-HNF1A for 16, 24 and 36 h. Flow-cytometry analyses of apoptosis activation (Annexin V staining) and cell size (forward light scatter) were performed simultaneously (Fig. 5). Annexin V detects the exposure of phosphatidylserine on the outer plasma membrane, a hallmark of apoptosis [28]. In agreement with our previous data [12, 13], we detected elevated levels of Annexin V-positive apoptotic cells from 36 h onwards (Fig. 5a–c). Control experiments using the general apoptosis activator staurosporine demonstrated that apoptosis induction was associated with a decrease in cell size, as cell shrinkage is a morphological hallmark of apoptosis (Fig. 5d). Therefore, only Annexin V-negative cells were evaluated for the analysis of cell-size alterations in response to DN-HNF1A. Quantification of the forward light-scatter signal in the non-apoptotic Annexin V-negative population indeed demonstrated a significant decrease in cell size after 24 and 36 h of DN-HNF1A expression (Fig. 5e).
Flow-cytometry analysis of cell size following DN-HNF1A induction. INS-1 cells were induced to overexpress DN-HNF1A (a non-induced; b induced 24 h; c induced 36 h; d STS 18 h) and were then stained with Annexin V and analysed by flow cytometry using the forward scatter channel. a–d The blue population represents cells staining positive for Annexin V, an indicator of apoptosis, while cells in red represent the non-apoptotic, Annexin V-negative population that was used for cell-size quantification. e Statistical analysis of the time-dependent decrease in cell size. Data are means + SD; *p < 0.05, difference from non-induced controls. A.U., arbitrary units; STS, staurosporine
Induction of DN-HNF1A is associated with an increase in p27 mRNA and protein levels
Inhibition of mTORC1 signalling leads to the hypophosphorylation of 4E-BP1 and enhanced binding to eIF4E, blocking cap-dependent translation and causing cell-cycle arrest at G1 [29]. Most eukaryotic mRNAs are translated through a cap-dependent mechanism of initiation. However, under conditions of cellular stress when cap-dependent translation is inhibited, such as nutrient deprivation and hypoxia, certain mRNAs can still be translated. Such conditions favour cap-independent translation of mRNAs containing an internal ribosome entry site (IRES) element within their 5 untranslated region [30]. p27 is a fundamental regulator of proliferation in most cell types that acts by blocking G1/S transition through inhibition of cyclin-dependent kinase 2 (CDK2) [31]. p27 mRNA stabilisation and a switch from cap-dependent to cap-independent p27 mRNA translation have been identified as key pathways leading to cell-cycle arrest upon inhibition of mTORC1 [32]. We determined mRNA expression of p27 in INS-1 cells induced to express DN-HNF1A by qPCR, and could detect a significant accumulation of p27 mRNA 16 h after induction (Fig. 6a). We furthermore observed an early (12 h) upregulation of p27 protein levels after the induction of DN-HNF1A by western blot analysis (Fig. 6b), suggesting that the induction of DN-HNF1A establishes conditions that favour an early accumulation of the cell-cycle inhibitor p27 in INS-1 cells.
Reduction in mTORC1 signalling is associated with increased mRNA and protein levels of the cell-cycle inhibitor p27. INS-1 cells were induced to overproduce DN-HNF1A for the indicated time periods. a Quantification of p27 mRNA using real-time quantitative PCR. Data shown are the means of n = 3 separate treatments. The experiment was repeated three times with similar results. *p < 0.05, difference from non-induced controls. b The expression of p27 was detected by western blotting using a p27-specific antibody. A duplicate experiment yielded similar results
Conclusion
Our study demonstrates that signalling through mTOR and specifically through the mTORC1 pathway is disturbed early during the suppression of HNF1A function in INS-1 insulinoma cells. mTOR activity is regulated by: the availability of growth factors, such as insulin and IGF-1; the accessibility of nutrients, notably the supply of key amino acids; and AMP levels [20, 33]. The role of mTOR is to integrate these diverse signals and to respond with alterations in cell metabolism, growth, proliferation, autophagy and survival [18]. Previous studies, and this study from our laboratory, have shown that prolonged, dominant negative suppression of HNF1A function causes a potent reduction in mTORC-2-dependent Ser473 phosphorylation of AKT1which coincided with the time points of maximal apoptosis [12]. Furthermore, overexpression of a constitutively active form of AKT1 was sufficient to rescue INS-1 cells from DN-HNF1A-induced apoptosis [12]. Hence, mTORC2 and AKT1 inactivation appear to mediate aspects of long-term alterations such as apoptosis activation in response to DN-HNF1A.
In the present study, we noted an early inactivation of mTORC1 signalling in response to DN-HNF1A expression, as demonstrated by the inhibition of Thr389 phosphorylation of S6K1 and Ser65 phosphorylation of 4E-BP1. mTORC1 signalling via phosphorylation and activation of S6K is an important regulator of cell size [18]. S6K1 phosphorylates the ribosomal protein S6 leading to selective translation of 5′ terminal oligopyrimidine tract (TOP) mRNAs. These oligopyrimidine-containing mRNAs encode for components of the translational apparatus including ribosomal proteins and translation elongation factors, thereby enhancing the translational capacity. It is interesting to mention in this context that a recent microarray analysis has also shown that DN-HNF1A expression decreases the transcription of several ribosomal proteins [34]. Targets of S6K1 other than ribosomal protein S6 have been identified, such as the transcription factor cAMP responsive element modulator (CREM)-τ, the RNA splicing/export factor cap binding protein 80 (CBP80) and the eukaryotic translation elongation factor 2 (eEF2) [35], which may mediate additional activities of S6K1 that were hitherto partially unknown. Loss of S6k1 in Drosophila is semi-lethal, and a markedly reduced body size was seen in the surviving adults [36]. This reduction in body size was caused by a decrease in cell size rather than a decrease in the actual cell number [36]. In mice, deletion of S6k1 was not shown to be lethal; however, the S6k1-deficient animals had a significantly reduced birthweight. Interestingly, the mice also had reduced circulating insulin levels resulting from a reduction in beta cell size [37]. In our studies we observed a time-dependent reduction in INS-1 cell size during dominant negative suppression of HNF1A function prior to the onset of apoptosis, thus suggesting that an early decrease in mTORC1 signalling leads to early reduction in cell size. A decrease in beta cell size (in addition to an overall decrease in beta cell mass) has also been suggested to occur in animal models of HNF1A-MODY [38]. It is tempting to speculate that such a decrease would also occur in HNF1A-MODY individuals. Since the insulin secretion rate is highly sensitive to alterations in the beta cell surface area [39], even small alterations in cell size could contribute to the reported insulin secretory defects in HNF1A-MODY [9].
mTOR has also been shown to hyperphosphorylate at several sites causing the liberation of eIF4E from 4E-BP and the association of eIF4E with both capped mRNA and eukaryotic-initiation-factor-4G (eIF4G). This leads to the assembly of the translation-initiation complex at the mRNA 5′ cap, promoting 5′ cap-dependent protein synthesis. A role for the switch from cap-dependent to cap-independent translation has been identified as a key signalling pathway in cell-cycle and apoptosis control [32, 40]. We could detect an early upregulation of p27 mRNA and protein levels in response to DN-HNF1A expression, prior to AKT1 inactivation [12]. Inhibition of mTORC1 has been shown to stabilise p27 mRNA and to promote cap-independent translation of p27 mRNA, hence both mRNA and protein levels may be upregulated independent of gene activation [31]. A recent study in human breast-cancer cell lines has shown that inhibition of mTORC1 with rapamycin also leads to reduced p27 protein degradation via decreased expression of the ubiquitin ligase subunit Skp2 [41]. Hence the early increase in p27 protein levels may also be due to a decrease in p27 degradation. However, we cannot fully exclude that p27 gene activation via AKT1 inactivation and subsequent forkhead box O3a (FOXO3a) activation may contribute to the early upregulation of p27 mRNA and protein levels observed in this and other studies. Together with previous reports demonstrating elevated p27 levels in cell culture and animal models of HNF1A-MODY [34, 42], our data therefore suggest that mTORC1 inhibition may also contribute to an early decrease in beta cell mass in HNF1A-MODY by inhibiting cell proliferation.
What could be the potential mechanisms responsible for the early mTORC1 vs late mTORC2 inactivation in response to DN-HNF1A? Our data demonstrate that gene expression changes of mTorc constituents appear to play no significant role in this differential inactivation. AKT1 activates mTORC1 and eventually S6K1 by a direct action on PRAS40 and via TSC1–TSC2 and RHEB GTPase [16]. Overexpression of a constitutively active form of Akt1 has been shown to be associated with an increased S6K activity [43]. It has also been shown that targeted overexpression of constitutively active Akt1 in transgenic mice induced a striking increase in beta cell mass caused by an increase in both beta cell number and size [44]. However, our study demonstrated different dephosphorylation kinetics in response to DN-HNF1A expression, suggesting that AKT1 inactivation may not be the primary mechanism of mTORC1 and S6K1 inactivation in this model. Importantly, cellular ATP depletion has also been reported to occur early during the induction of DN-HNF1A in INS-1 cells [22].
Energy depletion triggers the activation of AMP-activated protein kinase (AMPK) which, in turn, phosphorylates the TSC1–TSC2 complex leading to an inhibition of mTORC1[15]. We report here that although the basal phosphorylation of S6K1 was significantly reduced after induction of DN-HNF1A, mTORC1-dependent S6K1 phosphorylation was still partially responsive to high glucose. This glucose-stimulated activation of S6K1 is possibly mediated by increased ATP supply and reduced AMPK activation. Dominant-negative suppression of HNF1A has also been shown to reduce glucose-induced Ca2+ signalling in INS-1 cells [21], an event that may also contribute to the early inactivation of S6K1 and its partial rescue by high glucose, as increased intracellular Ca2+ concentrations have been shown to activate S6K1 but not AKT1 [45]. MAP kinase signalling pathways in particular are activated by increasing glucose and Ca2+ concentrations [46, 47]. A variety of proline-directed kinases including extracellular-signal-regulated kinase (ERK)1/2 and cell division cycle 2, G1 to S and G2 to M (CDC2) are capable of phosphorylating S6K1 in the C-terminal autoinhibitory pseudosubstrate domain of these sites in vitro [48], allowing control of the activation of S6K1 independently of the PI3K/PDK1 pathway. While multiple signalling pathways can act upon mTORC1 to regulate signalling through this complex, the mTORC2-signalling complex appears to be a more robust biological system. Indeed, mTORC1 appears to predominantly mediate short-term adaption to changes such as growth-factor supply, nutrient supply, hypoxia or stress, while mTORC2 is concerned with long-term changes such as spatial organisation and apoptosis. In our model, glucose did not affect mTORC2, and mTORC2-dependent AKT1 phosphorylation was not reduced until 48 h, as previously reported [12]. Reduced expression of Ins1, IGF1 and Akt1 genes in response to DN-HNF1A [12, 21, 33] may eventually lead to an inability to sustain high levels of active AKT1.
In summary, our data demonstrate that mTOR signalling via mTORC1 is disturbed early during the suppression of HNF1A function, and suggest that decreased signalling through mTORC1 may contribute to a disturbance of cell size and cell-cycle regulation in HNFF1A-MODY.
Abbreviations
- 4E-BP1:
-
eukaryotic translation initiation factor 4E binding protein 1
- AKT1:
-
thymoma viral proto-oncogene 1
- DN:
-
dominant-negative
- EIF4E:
-
eukaryotic translation initiation factor 4E
- HNF1A:
-
hepatocyte nuclear factor-1α
- INS-1:
-
insulinoma cell line
- GBL:
-
G protein β subunit-like
- mTOR:
-
mammalian target of rapamycin
- mTORC1:
-
mammalian target of rapamycin complex 1
- mTORC2:
-
mammalian target of rapamycin complex 2
- p27:
-
cyclin-dependent kinase inhibitor 1B
- PDK1:
-
phosphoinositide-dependent kinase-1
- PI3K:
-
phosphatidylinositol 3-kinase
- PRAS40:
-
proline-rich Akt substrate of 40 kDa
- qPCR:
-
real-time quantitative PCR
- Raptor:
-
regulator-associated protein of mTOR
- RHEB:
-
RAS-homologue enriched in brain
- Rictor:
-
rapamycin-insensitive companion of mTOR
- S6K1:
-
ribosomal protein S6 kinase
- SIN1:
-
stress-activated map kinase-interacting protein 1
- TSC1:
-
tuberous sclerosis 1 protein
- TSC2:
-
tuberous sclerosis 2 protein
- WT:
-
wild type
References
Fajans SS, Bell GI, Polonsky KS (2001) Molecular mechanisms and clinical pathophysiology of maturity-onset diabetes of the young. N Engl J Med 345:971–980
Frayling TM, Bulamn MP, Ellard S et al (1997) Mutations in the hepatocyte nuclear factor-1alpha gene are a common cause of maturity-onset diabetes of the young in the U.K. Diabetes 46:720–725
Ellard S, Colclough K (2006) Mutations in the genes encoding the transcription factors hepatocyte nuclear factor 1 alpha (HNF1A) and 4 alpha (HNF4A) in maturity-onset diabetes of the young. Human Mutat 27:854–869
Yamagata K, Oda N, Kaisaki PJ et al (1996) Mutations in the hepatocyte nuclear factor-1alpha gene in maturity-onset diabetes of the young (MODY3). Nature 384:455–458
Blumenfeld M, Maury M, Chouard T, Yaniv M, Condamine H (1991) Hepatic nuclear factor 1 (HNF1) shows a wider distribution than products of its known target genes in developing mouse. Development 113:589–599
Odom DT, Zizlsperger N, Gordon DB et al (2004) Control of pancreas and liver gene expression by HNF transcription factors. Science 303:1378–1381
Holmkvist J, Cervin C, Lyssenko V et al (2006) Common variants in HNF-1 alpha and risk of type 2 diabetes. Diabetologia 49:2882–2891
Pontoglio M, Sreenan S, Roe M et al (1998) Defective insulin secretion in hepatocyte nuclear factor 1alpha-deficient mice. J Clin Invest 101:2215–2222
Byrne MM, Sturis J, Menzel S et al (1996) Altered insulin secretory responses to glucose in diabetic and nondiabetic subjects with mutations in the diabetes susceptibility gene MODY3 on chromosome 12. Diabetes 45:1503–1510
Wang H, Hagenfeldt-Johansson K, Otten LA, Gauthier BR, Herrera PL, Wollheim CB (2002) Experimental models of transcription factor-associated maturity-onset diabetes of the young. Diabetes 51(Suppl 3):S333–S342
Wang H, Antinozzi PA, Hagenfeldt KA, Maechler P, Wollheim CB (2000) Molecular targets of a human HNF1 alpha mutation responsible for pancreatic beta-cell dysfunction. EMBO J 19:4257–4264
Wobser H, Bonner C, Nolan JJ, Byrne MM, Prehn JH (2006) Downregulation of protein kinase B/Akt-1 mediates INS-1 insulinoma cell apoptosis induced by dominant-negative suppression of hepatocyte nuclear factor-1alpha function. Diabetologia 49:519–526
Wobser H, Dussmann H, Kogel D et al (2002) Dominant-negative suppression of HNF-1 alpha results in mitochondrial dysfunction, INS-1 cell apoptosis, and increased sensitivity to ceramide-, but not to high glucose-induced cell death. J Biol Chem 277:6413–6421
Heitman J, Movva NR, Hall MN (1991) Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 253:905–909
Inoki K, Zhu T, Guan KL (2003) TSC2 mediates cellular energy response to control cell growth and survival. Cell 115:577–590
Sancak Y, Thoreen CC, Peterson TR et al (2007) PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol Cell 25:903–915
Guertin DA, Sabatini DM (2007) Defining the role of mTOR in cancer. Cancer Cell 12:9–22
Wullschleger S, Loewith R, Hall MN (2006) TOR signaling in growth and metabolism. Cell 124:471–484
Jacinto E, Facchinetti V, Liu D et al (2006) SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell 127:125–137
Sarbassov DD, Guertin DA, Ali SM, Sabatini DM (2005) Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307:1098–1101
Huang J, Dibble CC, Matsuzaki M, Manning BD (2008) The TSC1-TSC2 complex is required for proper activation of mTOR complex 2. Mol Cell Biol 28:4104–4115
Wang H, Maechler P, Hagenfeldt KA, Wollheim CB (1998) Dominant-negative suppression of HNF-1alpha function results in defective insulin gene transcription and impaired metabolism-secretion coupling in a pancreatic beta-cell line. EMBO J 17:6701–6713
Nicosia A, Monaci P, Tomei L et al (1990) A myosin-like dimerization helix and an extra-large homeodomain are essential elements of the tripartite DNA binding structure of LFB1. Cell 61:1225–1236
Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386
Gingras AC, Raught B, Sonenberg N (2001) Regulation of translation initiation by FRAP/mTOR. Genes Dev 15:807–826
Holz MK, Blenis J (2005) Identification of S6 kinase 1 as a novel mammalian target of rapamycin (mTOR)-phosphorylating kinase. J Biol Chem 280:26089–26093
Ruvinsky I, Meyuhas O (2006) Ribosomal protein S6 phosphorylation: from protein synthesis to cell size. Trends Biochem Sci 31:342–348
van Engeland M, Nieland LJ, Ramaekers FC, Schutte B, Reutelingsperger CP (1998) Annexin V-affinity assay: a review on an apoptosis detection system based on phosphatidylserine exposure. Cytometry 31:1–9
Wang X, Beugnet A, Murakami M, Yamanaka S, Proud CG (2005) Distinct signaling events downstream of mTOR cooperate to mediate the effects of amino acids and insulin on initiation factor 4E-binding proteins. Mol Cell Biol 25:2558–2572
Pause A, Belsham GJ, Gingras AC et al (1994) Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature 371:762–767
Sherr CJ, Roberts JM (1999) CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev 13:1501–1512
Miskimins WK, Wang G, Hawkinson M, Miskimins R (2001) Control of cyclin-dependent kinase inhibitor p27 expression by cap-independent translation. Mol Cell Biol 21:4960–4967
Gwinn DM, Shackelford DB, Egan DF et al (2008) AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell 30:214
Yang Q, Yamagata K, Fukui K et al (2002) Hepatocyte nuclear factor-1alpha modulates pancreatic beta-cell growth by regulating the expression of insulin-like growth factor-1 in INS-1 cells. Diabetes 51:1785–1792
Hannan KM, Brandenburger Y, Jenkins A et al (2003) mTOR-dependent regulation of ribosomal gene transcription requires S6K1 and is mediated by phosphorylation of the carboxy-terminal activation domain of the nucleolar transcription factor UBF. Mol Cell Biol 23:8862–8877
Montagne J, Stewart MJ, Stocker H, Hafen E, Kozma SC, Thomas G (1999) Drosophila S6 kinase: a regulator of cell size. Science 285:2126–2129
Ruvinsky I, Sharon N, Lerer T et al (2005) Ribosomal protein S6 phosphorylation is a determinant of cell size and glucose homeostasis. Genes Dev 19:2199–2211
Hagenfeldt-Johansson KA, Herrera PL, Wang H, Gjinovci A, Ishihara H, Wollheim CB (2001) Beta-cell-targeted expression of a dominant-negative hepatocyte nuclear factor-1 alpha induces a maturity-onset diabetes of the young (MODY)3-like phenotype in transgenic mice. Endocrinology 142:5311–5320
Giordano E, Cirulli V, Bosco D, Rouiller D, Halban P, Meda P (1993) B-cell size influences glucose-stimulated insulin secretion. Am J Physiol 265:C358–364
Luo Y, Marx SO, Kiyokawa H, Koff A, Massague J, Marks AR (1996) Rapamycin resistance tied to defective regulation of p27Kip1. Mol Cell Biol 16:6744–6751
Shapira M, Kakiashvili E, Rosenberg T, Hershko DD (2006) The mTOR inhibitor rapamycin down-regulates the expression of the ubiquitin ligase subunit Skp2 in breast cancer cells. Breast Cancer Res 8:R46
Luco RF, Maestro MA, del Pozo N, Philbrick WM, de la Ossa PP, Ferrer J (2006) A conditional model reveals that induction of hepatocyte nuclear factor-1alpha in Hnf1alpha-null mutant beta-cells can activate silenced genes postnatally, whereas overexpression is deleterious. Diabetes 55:2202–2211
Shioi T, McMullen JR, Kang PM et al (2002) Akt/protein kinase B promotes organ growth in transgenic mice. Mol Cell Biol 22:2799–2809
Fatrai S, Elghazi L, Balcazar N et al (2006) Akt induces beta-cell proliferation by regulating cyclin D1, cyclin D2, and p21 levels and cyclin-dependent kinase-4 activity. Diabetes 55:318–325
Conus NM, Hemmings BA, Pearson RB (1998) Differential regulation by calcium reveals distinct signaling requirements for the activation of Akt and p70S6k. J Biol Chem 273:4776–4782
Frodin M, Sekine N, Roche E et al (1995) Glucose, other secretagogues, and nerve growth factor stimulate mitogen-activated protein kinase in the insulin-secreting beta-cell line, INS-1. J Biol Chem 270:7882–7889
Khoo S, Cobb MH (1997) Activation of mitogen-activating protein kinase by glucose is not required for insulin secretion. Proc Natl Acad Sci U S A 94:5599–5604
Papst PJ, Sugiyama H, Nagasawa M, Lucas JJ, Maller JL, Terada N (1998) Cdc2-cyclin B phosphorylates p70 S6 kinase on Ser411 at mitosis. J Biol Chem 273:15077–15084
Acknowledgements
We thank H. Wang and C. Wollheim for the INS-1 clones and R. Cortese for kindly supplying the HNF1A antibody. This work was supported by a grant from the Health Research Board (RP/2004/220) to M. M. Byrne, and a grant from the Higher Education Authority (Program of Human Genomics) to J. H. M. Prehn.
Duality of interest
The authors declare that there is no duality of interest associated with this manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM
(PDF 74.0 KB)
ESM Table 1
Antibodies used in western blots (PDF 24.8 KB)
ESM Table 2
Rat primers for analysed genes (PDF 55.5 KB)
Rights and permissions
About this article
Cite this article
Farrelly, A.M., Wobser, H., Bonner, C. et al. Early loss of mammalian target of rapamycin complex 1 (mTORC1) signalling and reduction in cell size during dominant-negative suppression of hepatic nuclear factor 1-α (HNF1A) function in INS-1 insulinoma cells. Diabetologia 52, 136–144 (2009). https://doi.org/10.1007/s00125-008-1168-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-008-1168-8