Abstract
Aims/hypothesis
In order to identify type 2 diabetes disease susceptibility gene(s) in a Japanese population, we applied a region-wide case–control association test to the 20.4 Mb region between D3S1293 and D3S2319 on chromosome 3p24.3-22.1, supported by linkage to type 2 diabetes and its related traits in Japanese and multiple populations.
Materials and methods
We performed a two-stage association test using 1,762 Japanese persons with 485 gene-centric, evenly spaced, common single nucleotide polymorphism (SNP) markers with minor allele frequency >0.1. For mouse studies, total RNA was extracted from various organs of BKS.Cg-+Lepr db/+Lepr db and control mice, and from MIN6, NIH3T3 and C2C12 cell lines.
Results
We detected a landmark SNP375 (A/G) (rs2051211, p = 0.000046, odds ratio = 1.33, 95% CI 1.16–1.53) in intron 5 of the endonuclease G-like 1 (ENDOGL1) gene. Systematic dense SNPs approach identified a susceptibility linkage disequilibrium (LD) block of 116.5 kb by |D′|, an LD units map and a critical region of 2.1 kb by r 2 in ENDOGL1. A haplotype-based association test showed that an at-risk haplotype is associated with disease status (p = 0.00001). The expression of ENDOGL1 was rather ubiquitous with relatively abundant expression in the brain and also in a pancreatic islet beta cell line. Mouse Endogl1 expression increased in pancreatic islets of hyperglycaemic BKS.Cg-+Lepr db/+Lepr db mice compared with that in control mice.
Conclusions/interpretation
Based on the population genetics, fine mapping of LD block and haplotype analysis, we conclude that ENDOGL1 is a candidate disease-susceptibility gene for type 2 diabetes in a Japanese population. Further analysis in a larger sample size is required to substantiate this conclusion.
Similar content being viewed by others
Introduction
To identify the disease susceptibility gene(s) for type 2 diabetes, genetic association tests based on the candidate gene approach [1] or genome-wide association tests using single nucleotide polymorphisms (SNPs) [2–4] have been performed. We applied a region-wide association test, as an alternative and effective approach offering unbiased reliability, time and cost efficiency compared with a genome-wide association test. This was followed by SNPs-based fine-scale linkage disequilibrium (LD) and association mapping in the candidate region, supported by replicated linkage signals in Japanese and other multiple populations.
In a number of candidate regions [5–11] for type 2 diabetes and its related traits, we selected the overlapping region on chromosome 3p24.3-22.1. This region showed nominal significance in linkage (maximum logarithm of the odds [lod] score = 1.58) for type 2 diabetes in a Japanese population [12]. In Japanese people, it was reported to substantially overlap with that in Mexican Americans [6, 13] and with that reported in the American Diabetes Association’s Genetics of NIDDM study in Mexican Americans [14]. These results in Mexican Americans were replicated in Pima Indians [15] and Finnish families [16] (Electronic supplementary material [ESM] Table 1).
This candidate region, supported by replicated linkage signals including significant evidence of linkage with lod >3.6 [14], is expected to contain susceptibility gene(s). Studying this region, we focused on 44–70 cM in a Japanese population, assessing it here, in Japanese persons, by means of an association test. As selection criteria for the association test, we used gene-centric, evenly spaced common SNP markers, i.e. minor allele frequency (MAF) >0.1 at 10-kb intervals as far as possible. Our approach indicated that an association existed between multiple variants in the endonuclease G-like 1 (ENDOGL1) gene (MIM 604051) and type 2 diabetes.
Subjects and methods
Subjects with type 2 diabetes and control subjects
For this study, 1,762 Japanese subjects, consisting of 864 type 2 diabetic patients and 898 control subjects, were used. Type 2 diabetic patients were mainly recruited from the outpatient clinic of Tokushima University Hospital, Kyoto Prefectural University Hospital and their affiliated hospitals. The diagnosis of type 2 diabetes was based on the 1985 WHO criteria and patients were clinically defined as having gradual adult onset of the disease with medication. Patients with clinical criteria for monogenic forms of diabetes were excluded. Control subjects consisted of healthy adult members of the general population, who were intensively checked to ensure: (1) they had no family history of diabetes; (2) HbA1c levels were normal, i.e. <5.8%; (3) no other diseases were apparent on close clinical examination; and (4) that they were all of Japanese origin. Control subjects were mainly recruited from the Pharma SNP consortium (Web address, see ESM Table 2), a nationwide collection of Japanese control subjects obtained through the Health Science Research Resources Bank of the Japanese Collection of Research and Japan Health Science Foundation. The clinical characteristics of all subjects are presented in Table 1.
Genomic DNA was extracted from peripheral blood leucocytes or Epstein–Barr virus-immortalised B lymphocytes prepared by a standard protocol. We obtained informed consent from all subjects. All processes were approved by the Ethical Committee for Human Genome and Gene Research at the University of Tokushima in accordance with the tenets of the Declaration of Helsinki.
Animals/animal tissues
BKS.Cg-+Lepr db/+Lepr db (db mice) and BKS.Cg-m+/m+ (control mice) were purchased from CLEA Japan (Tokyo, Japan). Total RNA was extracted from the brain, liver, heart, kidney, lung, muscle, spleen, thymus, small intestine and pancreas in db and control mice, respectively, and from three cell lines (MIN6 pancreatic islet beta cell line and NIH3T3 and C2C12 non-beta cell lines) with Dneasy 96 (Qiagen, Stanford, CA, USA) and prepared with DNase treatment.
SNP markers and genotyping
We provisionally selected 539 gene-centric, evenly spaced, common SNP markers within the target region according to the following criteria: (1) suitability for designing optimal TaqMan high-throughput genotyping; (2) location in the gene region that we defined between 10 kb upstream of the transcription start site and 10 kb downstream of the final exon; (3) a distance of <10 kb between adjacent SNPs; and (4) common SNPs with a MAF >0.1 as our criteria for selecting SNPs for the association test from a prior selection of SNPs with a MAF >0.15 based on genotype data in 46 Japanese control individuals (24 males/22 females) (unpublished data supplied by Applied Biosystems, Foster City, CA, USA).
All SNP markers were genotyped by the TaqMan method according to the standard manufacturer’s protocol (TaqMan Universal PCR MasterMix, no uracil-N-glycosylase; Applied Biosystems). SNP markers that we were unable to amplify in this way were genotyped using QuantiTect Probe PCR Master Mix (Qiagen). Genotyping results were assessed by two researchers independently. To verify the genotyping accuracy, we genotyped 32 subjects for 30 randomly selected SNPs and retyped these subjects by direct sequencing analysis. Comparison of the results by two methods showed 100% concordance.
Study design of the association test
We conducted a two-stage case–control association test strategy in all samples by randomly assigning them to two independent panels (Fig. 1). In the first stage, we performed an association test of 310 type 2 diabetic patients and 361 control subjects (total: 671) at all SNP loci, and SNPs exhibiting significant allelic association (p < 0.05) were further examined in the second stage with independent replication in 554 type 2 diabetic patients and 537 control subjects (total: 1,091). We assessed the association by a χ2 test with a standard 2 × 2 allele frequency contingency table for the status diabetes or control. Using the PS program [17], we calculated the statistical power of this case-control test to detect association with a risk allele relating to MAF, a type 1 error rate and sample size. The false positive report probability (FPRP) was assessed by the method proposed by Wacholder et al. [18].
Analysis of stratification
We analysed the sub-structure of our samples by using the distribution of χ2 statistics for genome-wide unlinked SNP markers. We used GCONTROL software [19, 20] (Web address, see ESM Table 2) to assess the variability and magnitude of the test statistics observed at loci which are inflated in the event of population stratification or an unmeasured genetic relationship among the subjects. In subjects from the first stage in our study we analysed the genotype data of 92 SNP markers scattered throughout the genome with intervals of more than 1 Mb (SNP information available on request).
Analysis of linkage disequilibrium blocks
We assessed LD blocks in 310 type 2 diabetic and 361 control subjects by |D′| or r 2 > 0.9 according to Gabriel’s rule [21] using SNPAlyze_version 5.1 Pro (DYNACOM, Yokohama, Japan). We calculated LD units (LDU) to assess a recombination rate between LD blocks using LDMAP software [22]. The LDMAP is based on the Markov model and estimates the extent of LD between adjacent SNPs, which is expressed in LDU. To assess the LD block by LDU, we further assessed the local recombination rate between each pair of consecutive SNPs with PHASE version 2.2 software [23, 24]. A graphical overview of the |D′| or r 2 value was constructed using GOLD (Web address, see ESM Table 2). We analysed the haplotype with five SNPs in the r 2-based LD block using SNPAlyze, which is based on the expectation-maximisation algorithm.
Discovery of SNPs by sequencing around the ENDOGL1 gene
To discover novel SNPs in ENDOGL1, all six coding exons and 35 kb including intron 5 and 3′ downstream of ENDOGL1 were resequenced using the genomic DNA of 32 individuals (16 type 2 diabetic subjects, 16 control subjects), in addition to the search via the public database. All PCR products were prepared with ExoSAP-IT (Amersham Biosciences, Piscataway, NJ, USA) and sequenced using a terminator cycle sequencing kit (BigDye, version 1.1; Applied Biosystems) in both directions according to the standard manufacturer’s protocol and a 3730xl sequencer (Applied Biosystems).
Quantitative real time-PCR expression analysis of ENDOGL1
We performed quantitative RT-PCR analysis to estimate the expression of human ENDOGL1 or mouse Endogl1 using a 7900HT system with SYBR Green PCR Master Mix (Applied Biosystems). Human tissues were obtained from the adult human total RNA master panel (BD Bioscience, Palo Alto, CA, USA). RT-PCR conditions were based on the manufacturer’s protocols.
Electronic database information
The URLs for data used in this study are presented as ESM Table 2.
Results
Selection of SNPs in the target region
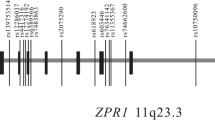
We selected the target region corresponding to a 20.4 Mb interval between D3S1293 (44.8 cM, map position [mp]: 21.9 Mb) and D3S2319 (70 cM, mp: 43.0 Mb) sequence-tagged-site markers. After quality control, we excluded 31 of the original 539 SNPs because they exhibited ambiguous genotyping qualities, leaving 508 SNPs for further study. Of these 508 SNPs, 499 were mapped within 80 genes (63% of the 128 genes based on NCBI Build 36.1 human genome assembly in the target region) (ESM Table 3) and nine were located in intergenic regions. After genotyping these SNPs, we selected 485 SNPs for further analysis. Our selection was based on Hardy–Weinberg equilibrium, p > 0.05 (χ 2 test), and MAF >0.1, because the power fell below 40% for SNPs with MAF < 0.1 and an odds ratio (OR) of 1.3. The average MAF of the 485 SNPs was 0.32 ± 0.003 in 310 type 2 diabetic subjects and 361 control subjects.
Association test in target region
In the first stage, a total of 485 SNPs was tested and analysed for association between SNP allele frequency and disease status. Of the 485 SNPs, 23 showed significant association with type 2 diabetes (p < 0.05) in the allele frequency (Fig. 2b, ESM Table 4). These 23 SNPs were analysed in the second stage following independent replication.
Results of association tests on the chromosome 3p24.3-22.1 region. The vertical bars a show gene position of the RefSeq genes and all selected SNPs markers. Allelic p values for allele frequency in association tests are shown: b in the first stage for 485 SNPs in 310 type 2 diabetic and 361 control subjects; c in the second stage for replication of 23 SNPs in 554 type 2 diabetic and 537 control subjects; and d in first and second stage subjects combined for 23 SNPs in 864 type 2 diabetic and 898 control subjects. p values are plotted against physical positions. The data denote the –log p. Bold line, p = 0.05; arrows, landmark SNP375 (rs2051211) located in ENDOGL1
Among these 23 SNPs, two of them, namely SNP158 (rs1857883) and SNP375 (rs2051211), showed a replicated association (p < 0.05) (Fig. 2c, ESM Table 4). Moreover, when all of the genotyping raw data of the first and second stages were combined, the landmark SNP375 in ENDOGL1 exhibited a peak of association in allele frequency with type 2 diabetes (p = 0.000046, OR = 1.33, 95% CI = 1.17–1.53) (Fig. 2d, ESM Table 4). SNP375 showed a significant association even after a conservative correction for multiple testing (Bonferroni-corrected p = 0.000046 × 485 = 0.022). The association between SNP375 and type 2 diabetes was confirmed to be independent of age and sex in the logistic regression analysis (data not shown). The genotype of SNP375, calculated by a χ2 test, showed statistical significance (p = 0.00016) (ESM Table 5). SNP158 also showed a significant association in the combined test (p = 0.00034, OR = 0.78, 95% CI = 0.69–0.90), but it was not positive after Bonferroni’s correction for multiple testing.
To exclude the possibility that our significant p values were false positives, we applied the FPRP method. To determine FPRP values, we used our experimentally determined OR values and the corresponding 95% CIs for SNP375 and SNP158 in the combined test. For SNP375 the FPRP value for a prior probability of 0.1% was 0.065 (0.413 for SNP158), satisfying a stringent FPRP value of <0.2 [18] and indicating that our findings were noteworthy (ESM Table 6). As a result, we postulated that SNP375 is a putative disease susceptibility SNP for type 2 diabetes in a Japanese population.
Detection power and assessment of population stratification
Power calculation showed that our association test with a sample size of 864 type 2 diabetic subjects and 898 control subjects affords a mean power >75% to detect the effect size of OR 1.3, a MAF of 0.3 and type 1 error rate of 0.05 (ESM Fig. 1a–c).
Using the Genomic Control method in the GCONTROL program, we assessed whether adverse stratification exists in our samples. Our results ruled out any harmful stratification in our population based on the inflation factor (data not shown). Significant allelic association of SNP375 was observed even after the GCONTROL program had corrected for latent population substructures (posterior probability = 0.940).
Determination of LD block structure around the landmark SNP375 and comprehensive detection of novel SNPs
To investigate the fine LD structure around SNP375 and the pattern of LD across the entire ENDOGL1 gene, we searched the public databases and found 14 SNPs (A-01; rs196377 to A-14; rs9873213) (ESM Table 7). Of these 14 SNPs, 9 (A-01 to A-09; rs2300668) were located 19.1–141 kb upstream from SNP375 and 5 (A-10; rs6810361 to A-14) were located 15.2–90.1 kb downstream from SNP375. To discover new SNPs, we resequenced the interval spanning 35 kb on both sides of SNP375 with a high LD coefficient (|D′| = 1.0). Genomic DNA from 32 control individuals was chosen on the basis of distribution of haplotype A-01/SNP375/SNP381 in an attempt to increase the probability of finding polymorphisms. Altogether we identified 31 SNPs, including five new SNPs; however, ten of the 31 SNPs were not common (MAF < 0.1). Consequently, we genotyped 35 SNPs (14 of A-01 to A-14 and 21 common SNPs from the 31 identified) with MAF >0.1 based on the same criteria for selection of SNPs for the association test. For our dense-SNPs typing, the |D′|-based type 2 diabetes susceptibility LD block was defined between A-02 (rs6807940) and B-19 (rs7647657), and included 40 SNPs that spanned 116.5 kb (Fig. 3, ESM Fig. 2).
LD block in the candidate region and p values around the landmark SNP375. Changes in LDU and recombination rates in the candidate region including the landmark SNP375 (rs2051211) (vertical arrow) were plotted a against physical positions of SNPs by LDMAP and PHASE. Bold line: estimates of changes in LDU by LDMAP; bold dotted line: estimates of recombination rate by PHASE version 2.0. Horizontal columns a: hatched, resequenced region; open, |D′|-based LD block; filled, r 2-based LD block. b Allelic p values for 45 SNPs between A-01 and A-11 in the ENDOGL1 and ACVR2B region are plotted against their physical positions. Bold line: p = 0.05
To further confirm the LD block structure, we examined LDU. We detected one main LD block in the association region that included SNPs between A-02 and B-19 and showed a low recombination rate. The LD block pattern in the LDU map was consistent with the recombination rate as estimated using PHASE software (Fig. 3a, the broken line). In ENDOGL1, the r 2-based tight LD block of 2.1 kb including significant SNPs was detected (Fig. 3a, ESM Fig. 3).
We also analysed the association of 40 SNPs in the |D′|-based LD block by comparing the allele frequency in all subjects. Our additional genotype analysis indicated that the following seven SNPs are significant: B-05 (rs6599210; mp 1.5 kb upstream from SNP375, p = 0.000348); B-07 (rs6599211; mp 0.9 kb upstream from SNP375, p = 0.047); B-08 (rs17037804; mp 0.5 kb upstream from SNP375, p = 0.000038); B-11 (rs2070490; mp 6.3 kb downstream from SNP375, p = 0.0012); and B-17 (new SNP; mp 8.8 kb downstream from SNP375, p = 0.0062) (Fig. 3b, ESM Table 7). Two SNPs B-18 (rs7649984) and B-19 near the LD block boundary showed a lower level of significance (p = 0.047, 0.048, respectively). The SNP B-08 remained significant after a conservative correction for multiple testing (Bonferroni-corrected p = 0.018). The FPRP value for SNP B-08 for a prior probability of 0.1% was 0.038 (<0.2).
Haplotype analysis in the candidate LD block
To assess the association between disease status and haplotype in the LD block, we selected all five SNPs in the r 2-based LD block, including the most significant SNP, and analysed haplotypes. We observed five major haplotypes composed of five SNPs (B-05, B-06, B-07, B-08 and SNP375), which covered more than 95% of haplotypes in the LD block (Table 2). We found a significant association between disease status and an at-risk haplotype Hap 2 (G-T-G-G-G, permutation p = 0.00001) and a disease-protective haplotype Hap 3 (A-T-G-A-A, permutation p = 0.0008).
Expression of human
ENDOGL1 or mouse Endogl1 Human ENDOGL1 expression was ubiquitous at relatively high levels, especially in the brain (ESM Fig. 4a). Mouse Endogl1 expression was also high in the brain. We observed statistically significant differences in Endogl1 mRNA levels in skeletal muscle and islets of hyperglycaemic mice at 12 weeks of age compared with those of normoglycaemic mice (p < 0.01) (ESM Fig. 4b). MIN6 cells also expressed a significantly higher level of mouse Endogl1 than did NIH3T3 or C2C12 cells (p < 0.01) (ESM Fig. 4c). These findings suggest that expression of Endogl1 is increased in islets of diabetic mice.
Discussion
SNP-based LD mapping of disease susceptibility regions by region-wide association has been proposed for the identification of susceptibility gene(s) for common diseases such as type 2 diabetes and rheumatoid arthritis [25, 26]. In an indirect association test in a Japanese population, we hypothesised that we would be able to efficiently detect the disease susceptibility gene(s) by using evenly spaced, common SNPs at approximately 10 kb intervals in the target region of replicated linkage evidence. Because the tag-SNPs resource from the HapMap [27] was not available at the onset of this study, we used SNPs yielding priorities in location and allele frequencies, rather than tag-SNPs drawn from LD information with a high-density SNP map.
In this study, we selected a target region on chromosome 3p24.3-22.1 that corresponds to 20.4 Mb based on replicated linkage for type 2 diabetes or its related traits [6, 12–16]. We adopted a two-step association test using 1,762 Japanese subjects to reduce the time and cost of genotyping. With prior simulation, the power using common SNP markers with a MAF >0.1 in the gene region and a sample size of 864 type 2 diabetic subjects and matched controls was >75%—sufficient to detect the susceptibility variant(s). One weakness of our association test is the age difference between the type 2 diabetic and control subjects (63.2 vs 41.0). Despite this age difference, the stratification analysis showed an absence of harmful stratification in our samples.
Our association test strategy successfully identified a significant association for one landmark SNP among the 485 analysed, namely SNP375 (rs2051211), located in intron 5 of ENDOGL1 (NM_005107). This SNP showed a strong allelic association with type 2 diabetes even after conservative Bonferroni’s correction. Not only the allele type, but also the genotype of SNP375 showed statistical significance. This was supported by the result of FPRP values. In logistic regression analysis, the association with SNP375 remained statistically significant after adjustment for age and sex. Because ageing is an important factor for developing diabetes, we suggest that the result of our association test be confirmed in later studies using age-matched control subjects.
LD mapping of the candidate region is a crucial step in identification of candidate gene(s) for disease and at-risk or protective haplotypes [2, 28]. Such strategies have also been shown to be useful in identifying novel or complex disease-related genes [29, 30]. We constructed a dense LD map and identified one block of 116.5 kb with a high LD coefficient (|D′| > 0.9). The LDU scale shows a pattern of ‘plateaus’ corresponding to regions of strong LD and ‘steps’ corresponding to regions of recombination. This pattern is mostly in concordance with recombination rates as assessed by PHASE. This |D′|-based LD block included two genes: ENDOGL1 and activin A receptor, type IIB (ACVR2B). The SNPs in ACVR2B were not associated with type 2 diabetes in this study. In contrast, not only the landmark SNP375 revealed significant association with type 2 diabetes, but also subsequent analysis of multiple additional SNPs within ENDOGL1 including the most significant SNP B-08 (rs17037804).
The |D′|-based LD block of 116.5 kb included a 2.1-kb LD block as defined by r 2 (range >0.85) around landmark SNP375. A haplotype-based association test revealed a significant result for at-risk and protective haplotypes, which we confirmed by a permutation test. Comparable p values were observed in single SNP association as well as haplotype association. On the basis of these observations, our study suggests that the variants in ENDOGL1 seem to confer susceptibility to type 2 diabetes. No phenotypic differences between carriers of the risk genotype compared with non-carriers were observed in control and/or type 2 diabetic subjects. Until now, however, the exact causal connection between these intronic SNPs and the risk of type 2 diabetes is not known, and the mechanism by which these candidate variants confer disease susceptibility remains to be elucidated.
The ENDOGL1 gene consists of six exons spanning approximately 31 kb of genomic DNA, while the 1.5 kb ENDOGL1 transcript encodes a protein of 368 amino acids [31]. Human ENDOGL1 expression was high in the brain and relatively low in insulin target tissues such as liver. Mouse Endogl1 expression was high in the brain, and lower in muscle of db mice than in that of control mice. Interestingly, mouse Endogl1 expression was significantly higher in the pancreatic islets of hyperglycaemic db mice than in control mice and that in MIN6 cell was higher than in non-beta cell lines. Although these observations suggest that ENDOGL1 protein may play a functional role in pancreatic islet beta cells, further investigation is obviously required before a firm conclusion can be drawn.
ENDOGL1 is a member of the DNA/RNA endonuclease family, including human [32], murine [33] and bovine [34] endonuclease G (ENDOG). The ENDOGL1 sequence shows 38.2, 37.1 and 36.5% amino acid identity to human, murine and bovine ENDOG, respectively. ENDOG is a mitochondria-specific nuclease that translocates to the nucleus during apoptosis [35]. Further study on a possible role of ENDOGL1 in the apoptotic pathway, especially in islet beta cells, is also required.
In conclusion, we have identified SNPs in the ENDOGL1 gene that show a significant association with type 2 diabetes. This is the first study to propose that ENDOGL1 may be a candidate gene for conferring susceptibility to type 2 diabetes in a Japanese population. To determine the consistency of our observation, replication studies in other independent populations should be given high priority.
Abbreviations
- ACVR2B:
-
activin A receptor, type IIB
- ENDOG:
-
endonuclease G
- ENDOGL1:
-
endonuclease G-like 1
- FPRP:
-
false positive report probability
- LD:
-
linkage disequilibrium
- LDU:
-
linkage disequilibrium units
- lod:
-
logarithm of the odds
- MAF:
-
minor allele frequency
- mp:
-
map position
- OR:
-
odds ratio
- SNP:
-
single nucleotide polymorphism
References
Barroso I, Luan J, PS Middelberg R et al (2003) Candidate gene association study in type 2 diabetes indicates a role for genes involved in β-cell function as well as insulin action. PLOS Biol 1:41–55
Hirschhorn JN, Daly MJ (2005) Genome-wide association studies for common diseases and complex traits. Nat Rev, Genet 6:95–108
Wang WY, Barratt BJ, Clayton DG, Todd JA (2005) Genome-wide association studies: theoretical and practical concerns. Nat Rev, Genet 6:109–118
Tanaka N, Babazono T, Saito S et al (2003) Association of solute carrier family 12 (sodium/chloride) member 3 with diabetic nephropathy, identified by genome-wide analyses of single nucleotide polymorphism. Diabetes 52:2848–2853
Elbein SC, Hoffman MD, Teng K, Leppert MF, Hasstedt SJ (1999) A genome-wide search for type 2 diabetes susceptibility genes in Utah Caucasians. Diabetes 48:1175–1182
Hanis CL, Boerwinkle E, Chakraborty R et al (1996) A genome-wide search for human non-insulin-dependent (type 2) diabetes genes reveals a major susceptibility locus on chromosome 2. Nat Genet 13:161–166
Vionnet N, El Hani H, Dupont S et al (2000) Genomewide search for type 2 diabetes-susceptibility genes in French whites: evidence for a novel susceptibility locus for early-onset diabetes on chromosome 3q27-qter and independent replication of a type 2-diabetes locus on chromosome 1q21-q24. Am J Hum Genet 67:1470–1480
Pezzolesi MG, Nam M, Nagase T et al (2004) Examination of candidate chromosomal regions for type 2 diabetes reveals a susceptibility locus on human chromosome 8p23.1. Diabetes 53:486–491
Duggirala R, Blangero J, Almasy L et al (1999) Linkage of type 2 diabetes mellitus and of age at onset to a genetic location on chromosome 10q in Mexican Americans. Am J Hum Genet 64:1127–1140
Bektas A, Suprenant ME, Wogan LT et al (1999) Evidence of a novel type 2 diabetes locus 50 cM centromeric to NIDDM2 on chromosome 12q. Diabetes 48:2246–2251
Ghosh S, Watanabe RM, Hauser ER et al (1999) Type 2 diabetes: evidence for linkage on chromosome 20 in 716 Finnish affected sib pairs. Proc Natl Acad Sci U S A 96:2198–2203
Iwasaki N, Cox NJ, Wang YQ et al (2003) Mapping genes influencing type 2 diabetes risk and BMI in Japanese subjects. Diabetes 52:209–213
Bell GI, Cox NJ, Lindner T et al (1997) Genetics of NIDDM in the Mexican–Americans of Starr country, Texas: an update. Diabet Rev 5:277–283
Ehm MG, Karmoub MC, Sakul H et al (2000) Genomewide search for type 2 diabetes susceptibility genes in four American populations. Am J Hum Genet 66:1871–1881
Pratley PE, Thompson DB, Prochazka M et al (1998) An autosomal genomic scan for loci linked to prediabetic phenotypes in Pima Indians. J Clin Invest 101:1757–1764
Watanabe RM, Ghosh S, Langefeld CD et al (2000) The Finland–United States investigation of non-insulin-dependent diabetes mellitus genetics (FUSION) study. II. An autosomal genome scan for diabetes-related quantitative-trait loci. Am J Hum Genet 67:1186–1200
Iwane M, Palensky J, Plante K (1997) A user’s review of commercial sample size software for design of biomedical studies using survival data. Control Clin Trials 18:65–83
Wacholder S, Chanock S, Garcia-Closas M, Ei Ghormli L, Rothman N (2004) Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J Natl Cancer Inst 96:434–442
Devlin B, Roeder K, Bacanu SA (2001) Unbiased methods for population-based association studies. Genet Epidemiol 21:273–284
Freedman ML, Reich D, Penney KL et al (2004) Assessing the impact of population stratification on genetic association studies. Nat Genet 36:388–393
Gabriel SB, Schaffner SF, Nguyen H et al (2002) The structure of haplotype blocks in the human genome. Science 296:2225–2229
Maniatis N, Collins A, Xu CF et al (2002) The first linkage disequilibrium (LD) maps: delineation of hot and cold blocks by diplotype analysis. Proc Natl Acad Sci U S A 99:2228–2233
Stephens M, Smith NJ, Donnelly P (2001) A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 68:978–989
Stephens M, Donnelly P (2003) A comparison of Bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet 73:1162–1169
Kato H, Nomura K, Osabe D et al (2006) Association of single-nucleotide polymorphisms in the suppressor of cytokine signaling 2 (SOCS2) gene with type 2 diabetes in the Japanese. Genomics 87:446–458
Hamada D, Takata Y, Osabe D et al (2005) Association between single-nucleotide polymorphisms in the SEC8L1 gene, which encodes a subset of the exocyst complex, and rheumatoid arthritis in a Japanese population. Arthritis Rheum 52:1371–1380
International HapMap Consortium (2005) A haplotype map of the human genome. Nature 437:1299–1320
De La Vega FM, Isaac H, Collins A et al (2005) The linkage disequilibrium maps of three human chromosomes across four populations reflect their demographic history and a common underlying recombination pattern. Genome Res 15:454–462
Klein RJ, Zeiss C, Chew EY et al (2005) Complement factor H polymorphism in age-related macular degeneration. Science 308:385–389
Suzuki A, Yamada R, Chang X et al (2003) Functional haplotypes of PADI4, encoding citrullinating enzyme peptidylarginine deiminase 4, are associated with rheumatoid arthritis. Nat Genet 34:395–402
Daigo Y, Isomura M, Nishiwaki T et al (1999) Characterization of a 1200-kb genomic segment of chromosome 3p22-p21.3. DNA Res 6:37–44
Tiranti V, Rossi E, Ruiz-Carrillo A et al (1995) Chromosomal localization of mitochondrial transcription factor A (TCF6), single-stranded DNA-binding protein (SSBP), and endonuclease G (ENDOG), three human housekeeping genes involved in mitochondrial biogenesis. Genomics 25:559–564
Prats E, Noel M, Letournean J et al (1997) Characterization and expression of the mouse endonuclease G gene. DNA Cell Biol 16:1111–1122
Gerschenson M, Houmiel KL, Low RL (1995) Endonuclease G from mammalian nuclei is identical to the major endonuclease of mitochondria. Nucl Acid Res 23:88–97
Li LY, Luo X, Wang X (2001) Endonuclease G is an apoptotic DNase when released from mitochondria. Nature 412:95–99
Acknowledgements
We sincerely thank our patients and control volunteers for donating blood samples and participating in this study. This study was supported by a grant from the Ministry of Education, Science and Technology (Knowledge Cluster Initiative).
Duality of interest
The authors have no conflict of interest to disclose in relation to the findings contained in this manuscript. In particular, commercial interests of Fujitsu company are not related to the subject matter of our paper, and the three authors with this affiliation do not have interests in the company other than their status as employees.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Below is the image is a link to a high resolution version
Supplementary Fig. 1
Power calculation of this case-control study. a Sample size required to detect association. The number of the type 2 diabetic subjects is set to 864. This value is required to attain between 60 and 85% power to detect association with the odds ratio varying between 1.25 and 1.35 and with a type 1 error rate of 0.05. b Power of this case-control association test to detect association against the risk allele frequency with a type 1 error rate of 0.05. c Power of a case-control test to detect association against the type 1 error rate varying between 0.05 and 0.0001. Graphs plotted with the PS program (GIF 134 kb)
Supplementary Fig. 2
LD mapping based on |D′| in the susceptibility region including a landmark SNP375 in the ENDOGL1 gene. LD block structure following Gabriel’s definition is shown, along with ENDOGL1 and ACVR2B (top), along with an exon/intron map of ENDOGL1 and ACVR2B including SNPs location A 116.5-kb LD block between markers A-02 and B-19 contains the ENDOGL1 and ACVR2B genes. LD block structure was analysed using genotyping data from 671 subjects, including 310 cases and 361 controls (GIF 60 kb)
Supplementary Fig. 3
LD block defined either by |D′| (lower right) or r 2 (upper left) in the susceptibility region. Each LD block was plotted separately by GOLD with a score of 1 (red) and 0 (dark blue). All 50 SNPs are listed at the bottom. A landmark SNP375 (rs2051211) and most significant B-08 (rs17037804) are indicated in red (GIF 67 kb)
Supplementary Fig. 4
Expression of human ENDOGL1 and mouse Endogl1. a Expression levels of human ENDOGL1 in ten different human tissues. Lanes 1 to 10 denote: brain, liver, heart, kidney, lung, muscle, spleen, thymus, small intestine, and pancreas, respectively. b Expression levels of mouse Endogl1 in eight different mouse tissues. Lanes 1 to 8 denote: liver, heart, adipose, muscle, lung, brain, kidney, and islet, respectively. Open columns, control mice (BKS.Cg-m+/m+) at 9 weeks of age; filled columns, diabetic mice (BKS.Cg-+Lepr db/+Lepr db; db mice) at 9 weeks of age with normal blood glucose concentrations after fasting for 16 h; hatched column (only lane 8), diabetic mice at 12 weeks of age with hyperglycaemia after fasting for 16 h (>13.8 mmol/l). c Expression levels of mouse Endogl1 in MIN6, NIH3T3 and C2C12. ** p < 0.01 for difference between db and control mice (b) and MIN6, NIH3T3 and C2C12 (c) as assessed by Student’s t test. Primer sequences: human ENDOGL1, sense primer sequence: 5′-GGAACAGAGGCAAGGTGTTACA-3′; antisense primer sequence: 5′-CTGCATCACCCATTATCTTGCT-3′; mouse Endogl1, sense primer sequence: 5′-GGAACAGAGACGAGGCGTTAC-3′; antisense primer sequence: 5′-TGTCAGCGTCACCGATGATCT-3′. Human ENDOGL1 or mouse Endogl1 gene expression was normalised for ENDOGL1 : β-actin ratio in five independent experiments using six mice in each group (GIF 56 kb)
Rights and permissions
About this article
Cite this article
Moritani, M., Nomura, K., Tanahashi, T. et al. Genetic association of single nucleotide polymorphisms in endonuclease G-like 1 gene with type 2 diabetes in a Japanese population. Diabetologia 50, 1218–1227 (2007). https://doi.org/10.1007/s00125-007-0631-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-007-0631-2