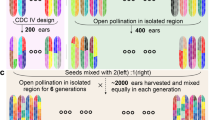
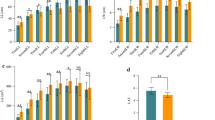
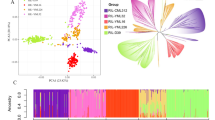
Abstract
Key message
A novel locus was discovered on chromosome 7 associated with a lesion mimic in maize; this lesion mimic had a quantitative and heritable phenotype and was predicted better via subset genomic markers than whole genome markers across diverse environments.
Abstract
Lesion mimics are a phenotype of leaf micro-spotting in maize (Zea mays L.), which can be early signs of biotic or abiotic stresses. Dissecting its inheritance is helpful to understand how these loci behave across different genetic backgrounds. Here, 538 maize recombinant inbred lines (RILs) segregating for a novel lesion mimic were quantitatively phenotyped in Georgia, Texas, and Wisconsin. These RILs were derived from three bi-parental crosses using a tropical pollinator (Tx773) as the common parent crossed with three inbreds (LH195, LH82, and PB80). While this lesion mimic was heritable across three environments based on phenotypic (\({H}_{\mathrm{p}}\) = 0.68) and genomic (\({H}_{\mathrm{g}}\) = 0.91) data, transgressive segregation was observed. A genome-wide association study identified a single novel locus on chromosome 7 (at 70.6 Mb) also covered by a quantitative trait locus interval (69.3–71.0 Mb), explaining 11–15% of the variation, depending on the environment. One candidate gene identified in this region, Zm00001eb308070, is related to the abscisic acid pathway involving in cell death. Genomic predictions were applied to genome-wide markers (39,611 markers) contrasted with a marker subset (51 markers). Population structure explained more variation than environment in genomic prediction, but other substantial genetic background effects were additionally detected. Subset markers explained substantially less genetic variation (24.9%) for the lesion mimic than whole genome markers (55.4%) in the model, yet predicted the lesion mimic better (0.56–0.66 vs. 0.26–0.29). These results indicate this lesion mimic phenotype was less affected by environment than by epistasis and genetic background effects, which explain its transgressive segregation.









Similar content being viewed by others
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Broman KW, Wu H, Sen Ś, Churchill GA (2003) R/qtl: QTL mapping in experimental crosses. Bioinformatics 19:889–890
Bruggeman Q, Prunier F, Mazubert C, De Bont L, Garmier M, Lugan R, Benhamed M, Bergounioux C, Raynaud C, Delarue M (2015) Involvement of arabidopsis hexokinase1 in cell death mediated by myo-inositol accumulation. Plant Cell 27:1801–1814
Crossa J, Montesinos-López OA, Pérez-Rodríguez P, Costa-Neto G, Fritsche-Neto R, Ortiz R, Martini JWR, Lillemo M, Montesinos-López A, Jarquin D, Breseghello F, Cuevas J, Rincent R (2022) Genome and environment based prediction models prediction models and methods of complex traits complex traits incorporating genotype × environment interaction. In: Ahmadi N, Bartholomé J (eds) Genomic prediction of complex traits: methods and protocols. Springer, New York, pp 245–283
DeKalb-Pfizer Genetics (1987) Corn 'PB80'. In: 008700174 PVPC (ed)
Furuta T, Ashikari M, Jena KK, Doi K, Reuscher S (2017) Adapting genotyping-by-sequencing for rice F2 populations. G3 Genes Genomes Genet 7:881–893
Gray J, Close PS, Briggs SP, Johal GS (1997) A Novel suppressor of cell death in plants encoded by the Lls1 gene of maize. Cell 89:25–31
Gray J, Janick-Buckner D, Buckner B, Close PS, Johal GS (2002) Light-dependent death of maize lls1 cells is mediated by mature chloroplasts. Plant Physiol 130:1894–1907
Holden's Foundation Seeds I (1985) Corn 'LH82'. In: Certificate PVP, 008500037 (eds)
Holden's Foundation Seeds I (1991) Corn 'LH195'. Plant variety protection certificate 009000047
Hu G, Richter TE, Hulbert SH, Pryor T (1996) Disease lesion mimicry caused by mutations in the rust resistance gene rp1. Plant Cell 8:1367–1376
Jambunathan N, Siani JM, McNellis TW (2001) A humidity-sensitive Arabidopsis copine mutant exhibits precocious cell death and increased disease resistance. Plant Cell 13:2225–2240
Johal GS, Hulbert SH, Briggs SP (1995) Disease lesion mimics of maize: a model for cell death in plants. BioEssays 17:685–692
Kosambi DD (2016) The estimation of map distances from recombination values. In: Ramaswamy R (ed) DD Kosambi: selected works in mathematics and statistics. Springer India, New Delhi, pp 125–130
Lorrain S, Vailleau F, Balagué C, Roby D (2003) Lesion mimic mutants: keys for deciphering cell death and defense pathways in plants? Trends Plant Sci 8:263–271
Mackay TF (2014) Epistasis and quantitative traits: using model organisms to study gene–gene interactions. Nat Rev Genet 15:22–33
Meng L, Li H, Zhang L, Wang J (2015) QTL IciMapping: integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J 3:269–283
Moeder W, Yoshioka K (2008) Lesion mimic mutants. Plant Signal Behav 3:764–767
Moore JH (2005) A global view of epistasis. Nat Genet 37:13–14
Mu X, Li J, Dai Z, Xu L, Fan T, Jing T, Chen M, Gou M (2021) Commonly and specifically activated defense responses in maize disease lesion mimic mutants revealed by integrated transcriptomics and metabolomics analysis. Front Plant Sci 12:690
Murray SC, Mayfield K, Pekar J, Brown P, Lorenz A, Isakeit T, Odvody G, Xu W, Betran J (2019) Tx741, Tx777, Tx779, Tx780, and Tx782 inbred maize lines for yield and southern united states stress adaptation. J Plant Regis 13:258–269
Neuffer MG, Calvert OH (1975) Dominant disease lesion mimics in maize. J Hered 66:265–270
Neuffer MG, Coe EH, Wessler SR (1997) Mutants of maize. Cold Spring Harbor Laboratory Press
Penning BW, Johal GS, McMullen MD (2004) A major suppressor of cell death, slm1, modifies the expression of the maize (Zea mays L.) lesion mimic mutation les23. Genome 47:961–969
Pérez P, de los Campos G (2014) Genome-wide regression and prediction with the BGLR statistical package. Genetics 198:483–495
Simmons C, Hantke S, Grant S, Johal GS, Briggs SP (1998) The maize lethal leaf spot 1 mutant has elevated resistance to fungal infection at the leaf epidermis. Mol Plant Microbe Interact 11:1110–1118
Smith SM, Steinau M, Trick HN, Hulbert SH (2010) Recombinant Rp1 genes confer necrotic or nonspecific resistance phenotypes. Mol Genet Genom 283:591–602
van Heerwaarden J, Hufford MB, Ross-Ibarra J (2012) Historical genomics of North American maize. Proc Natl Acad Sci 109:12420–12425
Walbot V (1991) Maize mutants for the 21st century. Plant Cell 3:851
Walbot V, Hoisington DA, Neuffer M (1983) Disease lesion mimic mutations. In: Genetic engineering of plants. Springer, pp 431–442
Wang S-H, Lim J-H, Kim S-S, Cho S-H, Yoo S-C, Koh H-J, Sakuraba Y, Paek N-C (2015) Mutation of SPOTTED LEAF3 (SPL3) impairs abscisic acid-responsive signalling and delays leaf senescence in rice. J Exp Bot 66:7045–7059
Zamir D (2001) Improving plant breeding with exotic genetic libraries. Nat Rev Genet 2:983–989
Funding
Texas Corn Producers Board supported this study. USDA-NIFA-AFRI project Award Nos. 2020-68013-32371 and 2021-67013-33915), Texas A&M AgriLife Research, USDA-NIFA-HATCH, the Eugene Butler Endowed Chair and USDA grant (1024073) in WI and GA. We appreciate the help of WI and GA crews as well as graduate students, staff, and undergraduate and high school employees of the Texas A&M Quantitative Genetics and Maize Breeding Program for their hard work and effort in maintaining this experiment, Dustin Eilert and Marina Borsecnik at University of Wisconsin, Madison, and Naomi Rodman at the University of Georgia.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection, genomic sequencing, growing materials in diverse environments and analysis were performed by AA, SCM, CIC, VI, JW, JIV, NS, TI, J-MA, JW, NDL, MAS, MB, JH, CDJ. The first draft of the manuscript was written by Alper Adak, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
There is no competing interests declared.
Ethics approval
This research did not use any research materials involving human or animal subjects.
Additional information
Communicated by Jiankang Wang.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Adak, A., Murray, S.C., Calderón, C.I. et al. Genetic mapping and prediction for novel lesion mimic in maize demonstrates quantitative effects from genetic background, environment and epistasis. Theor Appl Genet 136, 155 (2023). https://doi.org/10.1007/s00122-023-04394-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00122-023-04394-y