Abstract
Key message
Genetic models, QTLs and candidate gene for silique density on main inflorescence of rapeseed were identified.
Abstract
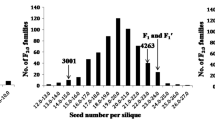
Silique density is one of the critical factors to determine seed yield and plant architecture in rapeseed (Brassica napus L.); however, the genetic control of this trait is largely unknown. In this study, the genetic model for silique density on main inflorescence (SDMI) of rapeseed was estimated according to the phenotypic data of P1 (an inbreed line with high SDMI), P2 (an inbreed line with low SDMI), F1, F2, BC1P1 and BC1P2 populations, revealing that SDMI is probably controlled by multi-minor genes with or without major gene. The QTLs for SDMI and its component characters including silique number on main inflorescence (SNMI) and main inflorescence length (MIL) were consequently mapped from a DH population derived from P1 and P2 by using a genetic linkage map constructed by restriction site-associated DNA sequencing (RAD seq) technology. A total of eight, 14 and three QTLs were identified for SDMI, SNMI and MIL under three environments, respectively, with an overlap among SDMI and SNMI in 55.7–75.4 cm on linkage group C06 which corresponding to 11.6–27.3 Mb on chromosome C06. Genomic resequencing was further conducted between a high- and a low-SDMI pool constructed from the DH population, and QTL-seq analysis identified a 0.15 Mb interval (25.98–26.13 Mb) from the C06-QTL region aforementioned. Transcriptome sequencing and qRT-PCR identified one possible candidate gene (BnARGOS) from the 0.15 Mb interval. This study will provide novel insights into the genetic basis of SD in rapeseed.



Similar content being viewed by others
Data availability
The plant material and datasets employed in this study are available from the corresponding author on reasonable request.
References
Audic S, Claverie J (1997) The significance of digital gene expression profiles. Genome Res 7:986–995
Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA, Selker EU, Cresko WA, Johnson EA (2008) Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE 3:e3376
Bin Y, Wei C, C. M, Fu T, Tu J, (2006) Mapping of quantitative trait loci for yield and yield components in Brassica napus L. Acta Agron Sin 32:676–682
Cai D, Xiao Y, Yang W, Ye W, Wang B, Younas M, Wu J, Liu K (2014) Association mapping of six yield-related traits in rapeseed (Brassica napus L.). Theor Appl Genet 127:85–96
Cai G, Yang Q, Chen H, Yang Q, Zhang C, Fan C, Zhou Y (2016) Genetic dissection of plant architecture and yield-related traits in Brassica napus. Scientific Reports 6:21625
Cao X, Liu B, Zhang Y (2013) SEA: a software package of segregation analysis of quantitative traits in plants. J Nanjing Agric Univ 36:1–6
Catchen JM, Amores A, Hohenlohe P, Cresko W, Postlethwait JH (2011) Stacks: building and genotyping loci de novo from short-read sequences. G3 (Bethesda) 1:171–182
Chandler J, Werr W (2017) DORNRÖSCHEN, DORNRÖSCHEN-LIKE, and PUCHI redundantly control floral meristem identity and organ initiation in Arabidopsis. J Exp Bot 68:3457–3472
Chen W, Zhang Y, Liu X, Chen B, Tu J, Fu T (2007) Detection of QTL for six yield-related traits in oilseed rape (Brassica napus) using DH and immortalized F2 populations. Theor Appl Genet 115:849–858
Cockerton HM, Vickerstaff RJ, Karlström A, Wilson F, Sobczyk M, He JQ, Sargent DJ, Passey AJ, Mcleary KJ, Pakozdi K (2018) Identification of powdery mildew resistance QTL in strawberry ( Fragaria × ananassa ). Theor Appl Genet 131:1995–2007
Dang Z, Zheng L, Wang J, Gao Z, Wu S, Qi Z, Wang Y (2013) Transcriptomic profiling of the salt-stress response in the wild recretohalophyte Reaumuria trigyna. BMC Genomics 14:29
Ding G, Zhao Z, Liao Y, Hu Y, Shi L, Long Y, Xu F (2012) Quantitative trait loci for seed yield and yield-related traits, and their responses to reduced phosphorus supply in Brassica napus. Ann Bot 109:747–759
Feng G, Qin Z, Yan J, Zhang X, Hu Y (2011) Arabidopsis ORGAN SIZE RELATED1 regulates organ growth and final organ size in orchestration with ARGOS and ARL. New Phytol 191:635–646
Hohenlohe PA, Bassham S, Etter PD, Stiffler N, Johnson EA, Cresko WA, Begun DJ (2010) Population genomics of parallel Adaptation in threespine stickleback using sequenced RAD tags. PLoS Genet 6:e1000862
Hu Y, Xie Q, Chua NH (2003) The Arabidopsis auxin-inducible gene ARGOS controls lateral organ size. Plant Cell 15:1951–1961
Hu Y, Poh HM, Chua NH (2006) The Arabidopsis ARGOS-LIKE gene regulates cell expansion during organ growth. Plant J 47:1–9
Huang Y, Shi J, Tao Z, Zhang L, Liu Q, Wang X, Yang Q, Liu G, Wang H, Zhang J (2014) Microarray expression analysis of the main inflorescence in Brassica napus. PLoS ONE 9:e102024
Institute SAS (2000) SAS/STAT user’s guide, version 8. SAS Institute, Cary
Mortazavi A, Williams B, Mccue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621–628
Özer H, Oral E, D. Ü, (1999) Relationships between yield and yield components on currently improved spring rapeseed cultivars. Turk J Agric for 23:603–608
Raboanatahiry N, Chao H, Hou D, Shi P, Li M (2018) QTL alignment for seed yield and yield related traits in Brassica napus. Front Plant Sci 9:1127
Radoev M, Becker HC, Ecke W (2008) Genetic analysis of heterosis for yield and yield components in rapeseed (Brassica napus L.) by quantitative trait locus mapping. Genetics 179:1547–1558
Ren Y, Cui C, Wang Q, Tang Z, Xu X, Lin N, Yin J, Li J, Zhou Q (2018) Genome-wide association analysis of silique density on racemes and its component traits in Brassica napus. Scientia Agricultura Sinica 51:1014–1033
Ross J, O’Neill D, Wolbang C, Symons G, Reid J (2001) Auxin-gibberell in interactions and their role in plant growth. J Plant Growth Regul 20:336–353
Shi J, Li R, Qiu D, Jiang C, Long Y, Morgan C, Bancroft I, Zhao J, Meng J (2009) Unraveling the complex trait of crop yield with quantitative trait loci mapping in Brassica napus. Genetics 182:851–861
Shi J, Zhan J, Yang Y, Ye J, Huang S, Li R, Wang X, Liu G, Wang H (2015) Linkage and regional association analysis reveal two new tightly-linked major-QTLs for pod number and seed number per pod in rapeseed (Brassica napus L.). Sci Rep 5:14481
Simard R, L’Ecuyer P (2011) Computing the two-sided Kolmogorov-Smirnov distribution. J Stat Softw 039:1–18
Takagi H, Abe A, Yoshida K, Kosugi S, Natsume S, Mitsuoka C, Uemura A, Utsushi H, Tamiru M, Takuno S (2013) QTL-seq: rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J 74:174–183
Tang M, Tong C, Liang L, Du C, Zhao J, Xiao L, Tong J, Dai X, Helal M, Dai W, Xiang Y (2020) A recessive high-density pod mutant resource of Brassica napus. Plant Sci 293:110411
Teo ZW, Song S, Wang YQ, Liu J, Yu H (2014) New insights into the regulation of inflorescence architecture. Trends Plant Sci 19:158–165
Tuncturk M, Ciftci V (2007) Relationship between yield and some yield components in rapeseed (Brassica napus ssp. oleifera L.) cultivars by using correlation and path analysis. Pak J Bot 39:81–84
Voorrips R (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78
Wang F, Guan C (2010) Molecular mapping and identification of quantitative trait loci for yield components in rapeseed (Brasscia napus L.). Hereditas 32:271–277
Wang X, Chen L, Wang A, Wang H, Tian J, Zhao X, Chao H, Zhao Y, Zhao W, Xiang J (2016) Quantitative trait loci analysis and genome-wide comparison for silique related traits in Brassica napus. BMC Plant Biol 16:71
Wang S, Basten C, Zeng Z (2011) Windows QTL Cartographer Version 2.5. Department of Statistics, North Carolina State University, Raleigh, NC
Wei C, Zhang Y, Liu X, Chen B, Tu J, Fu T (2007) Detection of QTL for six yield-related traits in oilseed rape (Brassica napus) using DH and immortalized F2 populations. Theor Appl Genet 115(6):849–858
Yang P, Shu C, Chen L, Xu J, Wu J, Liu K (2012) Identification of a major QTL for silique length and seed weight in oilseed rape (Brassica napus L.). Theor Appl Genet 125:285–296
Ye J, Yang Y, Chen B, Shi J, Luo M, Zhan J, Wang X, Liu G, Wang H (2017) An integrated analysis of QTL mapping and RNA sequencing provides further insights and promising candidates for pod number variation in rapeseed (Brassica napus L.). BMC Genomics 18:71
Yi Y, Shen Y, Li S, Ge X, Li Z (2017) High density linkage map construction and QTL detection for three silique-related traits in Orychophragmus violaceus derived Brassica napus population. Front Plant Sci 8:1512
Yu F, Shi X, Wang D (2014) Interpreting the genetic basis of silique traits in Brassica napus using a joint QTL network. Plant Breeding 133:52–60
Yuan D, Zhang Y, Wang Z, Qu C, Zhu D, Wan H, Liang Y (2022) BnKAT2 positively regulates the main inflorescence length and silique number in Brassica napus by regulating the auxin and cytokinin signaling pathways. Plants (basel) 11:1679
Zhang Y, Gai J (2000a) Identification of two major genes plus polygenes mixed inheritance model of quantitative traits in B1 and B2, and F2. J Biomath 15:358–366
Zhang Y, Gai J (2000b) The IECM algorithm for estimation of component distribution parameters in segregating analysis of quantitative traits. Acta Agron Sin 26:699–706
Zhang L, Yang G, Liu P, Hong D, Li S, He Q (2011) Genetic and correlation analysis of silique-traits in Brassica napus L. by quantitative trait locus mapping. Theor Appl Genet 122:21–31
Zhang L, Li S, Chen L, Yang G (2012) Identification and mapping of a major dominant quantitative trait locus controlling seeds per silique as a single Mendelian factor in Brassica napus L. Theor Appl Genet 125:695–705
Zhao W, Wang X, Wang H, Tian J, Li B, Chen L, Chao H, Long Y, Xiang J, Gan J (2016) Genome-wide identification of QTL for seed yield and yield-related traits and construction of a high-density consensus map for QTL comparison in Brassica napus. Front Plant Sci 7:17
Zhou Z, Jiang Y, Wang Z, Gou Z, Lyu J, Li W, Yu Y, Shu L, Zhao Y, Ma Y (2015) Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean. Nat Biotechnol 33:408–414
Acknowledgement
We thank for the financial support from the following findings.
Funding
This study was funded by the National Key R&D Program of China (2022ZD0400802) and Sichuan and Chongqing joint key R & D project (cstc2021jscx-cylhX0003).
Author information
Authors and Affiliations
Contributions
JM and WQ directed the experiment and revised the manuscript. XM and JW carried out the project and wrote the first draft of the manuscript. YG, PF, WN and RL conducted the phenotypic evaluation.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Additional information
Communicated by Benjamin Stich.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ma, X., Wang, J., Gu, Y. et al. Genetic analysis and QTL mapping for silique density in rapeseed (Brassica napus L.). Theor Appl Genet 136, 128 (2023). https://doi.org/10.1007/s00122-023-04375-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00122-023-04375-1