Abstract
Key message
The fiber length-related qFL-A12-5 identified in CSSLs introgressed from Gossypium barbadense into Gossypium hirsutum was fine-mapped to an 18.8 kb region on chromosome A12, leading to the identification of the GhTPR gene as a potential regulator of cotton fiber length.
Abstract
Fiber length is a key determinant of fiber quality in cotton, and it is a key target of artificial selection for breeding and domestication. Although many fiber length-related quantitative trait loci have been identified, there are few reports on their fine mapping or candidate gene validation, thus hampering efforts to understand the mechanistic basis of cotton fiber development. Our previous study identified the qFL-A12-5 associated with superior fiber quality on chromosome A12 in the chromosome segment substitution line (CSSL) MBI7747 (BC4F3:5). A single segment substitution line (CSSL-106) screened from BC6F2 was backcrossed to construct a larger segregation population with its recurrent parent CCRI45, thus enabling the fine mapping of 2852 BC7F2 individuals using denser simple sequence repeat markers to narrow the qFL-A12-5 to an 18.8 kb region of the genome, in which six annotated genes were identified in Gossypium hirsutum. Quantitative real-time PCR and comparative analyses led to the identification of GH_A12G2192 (GhTPR) encoding a tetratricopeptide repeat-like superfamily protein as a promising candidate gene for qFL-A12-5. A comparative analysis of the protein-coding regions of GhTPR among Hai1, MBI7747, and CCRI45 revealed two non-synonymous mutations. The overexpression of GhTPR resulted in longer roots in Arabidopsis, suggesting that GhTPR may regulate cotton fiber development. These results provide a foundation for future efforts to improve cotton fiber length.





Similar content being viewed by others
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.
References
Altschul S, Gish W, Miller W, Myers E, Lipman D (1990) Basic local alignment search tool. J Mol Biol 215(3):403–410. https://doi.org/10.1016/S0022-2836(05)80360-2
Cao JF, Zhao B, Huang CC, Chen ZW, Zhao T, Liu HR, Hu GJ, Shangguan XX, Shan CM, Wang LJ, Zhang TZ, Wendel JF, Guan XY, Chen XY (2020) The miR319-targeted GhTCP4 promotes the transition from cell elongation to wall thickening in cotton fiber. Mol Plant 13(7):1063–1077. https://doi.org/10.1016/j.molp.2020.05.006
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16(6):735–743. https://doi.org/10.1046/j.1365-313x.1998.00343.x
Fang DD, Naoumkina M, Thyssen GN, Bechere E, Li P, Florane CB (2020) An EMS-induced mutation in a tetratricopeptide repeat-like superfamily protein gene (Ghir_A12G008870) on chromosome A12 is responsible for the liy short fiber phenotype in cotton. Theor Appl Genet 133(1):271–282. https://doi.org/10.1007/s00122-019-03456-4
Fang L, Wang Q, Hu Y, Jia Y, Chen J, Liu B, Zhang Z, Guan X, Chen S, Zhou B, Mei G, Sun J, Pan Z, He S, Xiao S, Shi W, Gong W, Liu J, Ma J, Cai C, Zhu X, Guo W, Du X, Zhang T (2017a) Genomic analyses in cotton identify signatures of selection and loci associated with fiber quality and yield traits. Nat Genet 49(7):1089–1098. https://doi.org/10.1038/ng.3887
Fang X, Liu X, Wang X, Wang W, Liu D, Zhang J, Liu D, Teng Z, Tan Z, Liu F, Zhang F, Jiang M, Jia X, Zhong J, Yang J, Zhang Z (2017b) Fine-mapping qFS07.1 controlling fiber strength in upland cotton (Gossypium hirsutum L.). Theor Appl Genet 130(4):795–806. https://doi.org/10.1007/s00122-017-2852-1
Feng H, Li X, Chen H, Deng J, Zhang C, Liu J, Wang T, Zhang X, Dong J (2018) GhHUB2, a ubiquitin ligase, is involved in cotton fiber development via the ubiquitin-26S proteasome pathway. J Exp Bot 69(21):5059–5075. https://doi.org/10.1093/jxb/ery269
Feng L, Zhou C, Su Q, Xu M, Yue H, Zhang S, Zhou B (2020) Fine-mapping and candidate gene analysis of qFS-Chr. D02, a QTL for fibre strength introgressed from a semi-wild cotton into Gossypium hirsutum. Plant Sci 297:110524. https://doi.org/10.1016/j.plantsci.2020.110524
Forsthoefel NR, Klag KA, Simeles BP, Reiter R, Brougham L, Vernon DM (2013) The Arabidopsis Plant Intracellular Ras-group LRR (PIRL) family and the value of reverse genetic analysis for identifying genes that function in gametophyte development. Plants (basel) 2(3):507–520. https://doi.org/10.3390/plants2030507
Forsthoefel NR, Vernon DM (2011) Effect of sporophytic PIRL9 genotype on post-meiotic expression of the Arabidopsis pirl1;pirl9 mutant pollen phenotype. Planta 233(2):423–431. https://doi.org/10.1007/s00425-010-1324-5
Gaudet P, Livstone MS, Lewis SE, Thomas PD (2011) Phylogenetic-based propagation of functional annotations within the Gene Ontology consortium. Brief Bioinform 12(5):449–462. https://doi.org/10.1093/bib/bbr042
Guan X, Lee JJ, Pang M, Shi X, Stelly DM, Chen ZJ (2011) Activation of Arabidopsis seed hair development by cotton fiber-related genes. PLoS ONE 6(7):e21301. https://doi.org/10.1371/journal.pone.0021301
He S, Sun G, Geng X, Gong W, Dai P, Jia Y, Shi W, Pan Z, Wang J, Wang L, Xiao S, Chen B, Cui S, You C, Xie Z, Wang F, Sun J, Fu G, Peng Z, Hu D, Wang L, Pang B, Du X (2021) The genomic basis of geographic differentiation and fiber improvement in cultivated cotton. Nat Genet 53(6):916–924. https://doi.org/10.1038/s41588-021-00844-9
Hossain MF, Sultana MM, Tanaka A, Dutta AK, Hachiya T, Nakagawa T (2022) Expression analysis of plant intracellular Ras-group related leucine-rich repeat proteins (PIRLs) in Arabidopsis thaliana. Biochem Biophys Rep 30:101241. https://doi.org/10.1016/j.bbrep.2022.101241
Hu Y, Chen J, Fang L, Zhang Z, Ma W, Niu Y, Ju L, Deng J, Zhao T, Lian J, Baruch K, Fang D, Liu X, Ruan YL, Rahman MU, Han J, Wang K, Wang Q, Wu H, Mei G, Zang Y, Han Z, Xu C, Shen W, Yang D, Si Z, Dai F, Zou L, Huang F, Bai Y, Zhang Y, Brodt A, Ben-Hamo H, Zhu X, Zhou B, Guan X, Zhu S, Chen X, Zhang T (2019) Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat Genet 51(4):739–748. https://doi.org/10.1038/s41588-019-0371-5
Huang G, Huang JQ, Chen XY, Zhu YX (2021) Recent advances and future perspectives in cotton research. Annu Rev Plant Biol 72:437–462. https://doi.org/10.1146/annurev-arplant-080720-113241
Huang Y, Liu X, Tang K, Zuo K (2013) Functional analysis of the seed coat-specific gene GbMYB2 from cotton. Plant Physiol Biochem 73:16–22. https://doi.org/10.1016/j.plaphy.2013.08.004
Islam MS, Zeng L, Thyssen GN, Delhom CD, Kim HJ, Li P, Fang DD (2016) Mapping by sequencing in cotton (Gossypium hirsutum) line MD52ne identified candidate genes for fiber strength and its related quality attributes. Theor Appl Genet 129:1071–1086. https://doi.org/10.1007/s00122-016-2684-4
Jiang SY, Ramamoorthy R, Ramachandran S (2008) Comparative transcriptional profiling and evolutionary analysis of the GRAM domain family in eukaryotes. Dev Biol 314(2):418–432. https://doi.org/10.1016/j.ydbio.2007.11.031
Jiang Y, Guo W, Zhu H, Ruan YL, Zhang T (2012) Overexpression of GhSusA1 increases plant biomass and improves cotton fiber yield and quality. Plant Biotechnol J 10(3):301–312. https://doi.org/10.1111/j.1467-7652.2011.00662.x
Lange PR, Geserick C, Tischendorf G, Zrenner R (2008) Functions of chloroplastic adenylate kinases in Arabidopsis. Plant Physiol 146(2):492–504. https://doi.org/10.1104/pp.107.114702
Lei M, Li H, Zhang L, Wang J (2015) QTL IciMapping: integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. The Crop Journal 3:269–283. https://doi.org/10.1016/j.cj.2015.01.001
Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30(1):325–327. https://doi.org/10.1093/nar/30.1.325
Leskow CC, Kamenetzky L, Dominguez PG, Diaz Zirpolo JA, Obata T, Costa H, Marti M, Taboga O, Keurentjes J, Sulpice R, Ishihara H, Stitt M, Fernie AR, Carrari F (2016) Allelic differences in a vacuolar invertase affect Arabidopsis growth at early plant development. J Exp Bot 67(14):4091–4103. https://doi.org/10.1093/jxb/erw185
Li Q, Wan JM (2005) SSRHunter: development of a local searching software for SSR sites. Hereditas 27:808–810. https://doi.org/10.16288/j.yczz.2005.05.024
Li SQ, Liu AY, Kong LL, Gong JW, Li JW, Gong WK, Lu QW, Li PT, Ge Q, Shang HH, Xiao XH, Liu RX, Zhang Q, Shi YZ, Yuan YL (2019) QTL mapping and genetic effect of chromosome segment substitution lines with excellent fiber quality from Gossypium hirsutum × Gossypium barbadense. Mol Genet Genomics 294(5):1123–1136. https://doi.org/10.1007/s00438-019-01566-8
Li Z, Wang P, You C, Yu J, Zhang X, Yan F, Ye Z, Shen C, Li B, Guo K, Liu N, Thyssen GN, Fang DD, Lindsey K, Zhang X, Wang M, Tu L (2020) Combined GWAS and eQTL analysis uncovers a genetic regulatory network orchestrating the initiation of secondary cell wall development in cotton. New Phytol 226(6):1738–1752. https://doi.org/10.1111/nph.16468
Liu B, Zhu Y, Zhang T, Zhang X (2015) The R3-MYB Gene GhCPC negatively regulates cotton fiber elongation. PLoS ONE 10(2):e0116272. https://doi.org/10.1371/journal.pone.0116272
Liu R, Gong J, Xiao X, Zhang Z, Li J, Liu A, Lu Q, Shang H, Shi Y, Ge Q, Iqbal MS, Deng X, Li S, Pan J, Duan L, Zhang Q, Jiang X, Zou X, Hafeez A, Chen Q, Geng H, Gong W, Yuan Y (2018) GWAS analysis and QTL identification of fiber quality traits and yield components in upland cotton using enriched high-density SNP markers. Front Plant Sci 9:1067. https://doi.org/10.3389/fpls.2018.01067
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Lu Q, Xiao X, Gong J, Li P, Zhao Y, Feng J, Peng R, Shi Y, Yuan Y (2021) Identification of candidate cotton genes associated with fiber length through quantitative trait loci mapping and RNA-sequencing using a chromosome segment substitution line. Front Plant Sci 12:796722. https://doi.org/10.3389/fpls.2021.796722
Ma J, Jiang Y, Pei W, Wu M, Ma Q, Liu J, Song J, Jia B, Liu S, Wu J, Zhang J, Yu J (2022) Expressed genes and their new alleles identification during fibre elongation reveal the genetic factors underlying improvements of fibre length in cotton. Plant Biotechnol J. https://doi.org/10.1111/pbi.13874
Ma Z, He S, Wang X, Sun J, Zhang Y, Zhang G, Wu L, Li Z, Liu Z, Sun G, Yan Y, Jia Y, Yang J, Pan Z, Gu Q, Li X, Sun Z, Dai P, Liu Z, Gong W, Wu J, Wang M, Liu H, Feng K, Ke H, Wang J, Lan H, Wang G, Peng J, Wang N, Wang L, Pang B, Peng Z, Li R, Tian S, Du X (2018) Resequencing a core collection of upland cotton identifies genomic variation and loci influencing fiber quality and yield. Nat Genet 50(6):803–813. https://doi.org/10.1038/s41588-018-0119-7
Machado A, Wu Y, Yang Y, Llewellyn DJ, Dennis ES (2009) The MYB transcription factor GhMYB25 regulates early fibre and trichome development. Plant J 59(1):52–62. https://doi.org/10.1111/j.1365-313X.2009.03847.x
Naoumkina M, Thyssen GN, Fang DD, Jenkins JN, McCarty JC, Florane CB (2019) Genetic and transcriptomic dissection of the fiber length trait from a cotton (Gossypium hirsutum L.) MAGIC population. BMC Genomics 20(1):112. https://doi.org/10.1186/s12864-019-5427-5
Naoumkina M, Zeng L, Fang DD, Wang M, Thyssen GN, Florane CB, Li P, Delhom CD (2020) Mapping and validation of a fiber length QTL on chromosome D11 using two independent F2 populations of upland cotton. Mol Breeding 40:31. https://doi.org/10.1007/s11032-020-01111-1
Nie X, Wen T, Shao P, Tang B, Nuriman-Guli A, Yu Y, Du X, You C, Lin Z (2020) High-density genetic variation maps reveal the correlation between asymmetric interspecific introgressions and improvement of agronomic traits in Upland and Pima cotton varieties developed in Xinjiang. China Plant J 103(2):677–689. https://doi.org/10.1111/tpj.14760
Nunome T, Negoro S, Kono I, Kanamori H, Miyatake K, Yamaguchi H, Ohyama A, Fukuoka H (2009) Development of SSR markers derived from SSR-enriched genomic library of eggplant (Solanum melongena L.). Theor Appl Genet 119(6):1143–1153. https://doi.org/10.1007/s00122-009-1116-0
Ohtake Y, Wickner RB (1995) Yeast virus propagation depends critically on free 60S ribosomal subunit concentration. Mol Cell Biol 15(5):2772–2781. https://doi.org/10.1128/MCB.15.5.2772
Paterson AH, Wendel JF, Gundlach H, Guo H, Jenkins J, Jin D, Llewellyn D, Showmaker KC, Shu S, Udall J, Yoo MJ, Byers R, Chen W, Doron-Faigenboim A, Duke MV, Gong L, Grimwood J, Grover C, Grupp K, Hu G, Lee TH, Li J, Lin L, Liu T, Marler BS, Page JT, Roberts AW, Romanel E, Sanders WS, Szadkowski E, Tan X, Tang H, Xu C, Wang J, Wang Z, Zhang D, Zhang L, Ashrafi H, Bedon F, Bowers JE, Brubaker CL, Chee PW, Das S, Gingle AR, Haigler CH, Harker D, Hoffmann LV, Hovav R, Jones DC, Lemke C, Mansoor S, ur Rahman M, Rainville LN, Rambani A, Reddy UK, Rong JK, Saranga Y, Scheffler BE, Scheffler JA, Stelly DM, Triplett BA, Van Deynze A, Vaslin MF, Waghmare VN, Walford SA, Wright RJ, Zaki EA, Zhang T, Dennis ES, Mayer KF, Peterson DG, Rokhsar DS, Wang X, Schmutz J (2012) Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492(7429):423–427. https://doi.org/10.1038/nature11798
Pellett S, Tracy JW (2006) Mak16p is required for the maturation of 25S and 5.8S rRNAs in the yeast Saccharomyces cerevisiae. Yeast 23(7):495–506. https://doi.org/10.1002/yea.1368
Ren Z, Yu D, Yang Z, Li C, Qanmber G, Li Y, Li J, Liu Z, Lu L, Wang L, Zhang H, Chen Q, Li F, Yang Z (2017) Genome-wide identification of the MIKC-Type MADS-box gene family in Gossypium hirsutum L. unravels their roles in flowering. Front Plant Sci 8:384. https://doi.org/10.3389/fpls.2017.00384
Ruan YL, Chourey PS, Delmer DP, Perezgrau L (1997) The differential expression of sucrose synthase in relation to diverse patterns of carbon partitioning in developing cotton seed. Plant Physiol 115(2):375–385. https://doi.org/10.1104/pp.115.2.375
Ruan YL, Llewellyn DJ, Furbank RT (2003) Suppression of sucrose synthase gene expression represses cotton fiber cell initiation, elongation, and seed development. Plant Cell 15(4):952–964. https://doi.org/10.1105/tpc.010108
Salih H, Gong W, He S, Sun G, Sun J, Du X (2016) Genome-wide characterization and expression analysis of MYB transcription factors in Gossypium hirsutum. BMC Genet 17(1):129. https://doi.org/10.1186/s12863-016-0436-8
Schapire AL, Valpuesta V, Botella MA (2006) TPR proteins in plant hormone signaling. Plant Signal Behav 1(5):229–230. https://doi.org/10.4161/psb.1.5.3491
Serna L, Martin C (2006) Trichomes: different regulatory networks lead to convergent structures. Trends Plant Sci 11(6):274–280. https://doi.org/10.1016/j.tplants.2006.04.008
Shan CM, Shangguan XX, Zhao B, Zhang XF, Chao LM, Yang CQ, Wang LJ, Zhu HY, Zeng YD, Guo WZ, Zhou BL, Hu GJ, Guan XY, Chen ZJ, Wendel JF, Zhang TZ, Chen XY (2014) Control of cotton fibre elongation by a homeodomain transcription factor GhHOX3. Nat Commun 5:5519. https://doi.org/10.1038/ncomms6519
Shi Y, Li W, Li A, Ge R, Zhang B, Li J, Liu G, Li J, Liu A, Shang H, Gong J, Gong W, Yang Z, Tang F, Liu Z, Zhu W, Jiang J, Yu X, Wang T, Wang W, Chen T, Wang K, Zhang Z, Yuan Y (2015) Constructing a high-density linkage map for Gossypium hirsutum × Gossypium barbadense and identifying QTLs for lint percentage. J Integr Plant Biol 57(5):450–467. https://doi.org/10.1111/jipb.12288
Shi Y, Liu A, Li J, Zhang J, Li S, Zhang J, Ma L, He R, Song W, Guo L, Lu Q, Xiang X, Gong W, Gong J, Ge Q, Shang H, Deng X, Pan J, Yuan Y (2020) Examining two sets of introgression lines across multiple environments reveals background-independent and stably expressed quantitative trait loci of fiber quality in cotton. Theor Appl Genet 133(7):2075–2093. https://doi.org/10.1007/s00122-020-03578-0
Shi Y, Zhang B, Liu A, Li W, Li J, Lu Q, Zhang Z, Li S, Gong W, Shang H, Gong J, Chen T, Ge Q, Wang T, Zhu H, Liu Z, Yuan Y (2016) Quantitative trait loci analysis of Verticillium wilt resistance in interspecific backcross populations of Gossypium hirsutum × Gossypium barbadense. BMC Genomics 17(1):877. https://doi.org/10.1186/s12864-016-3128-x
Shi YH, Zhu SW, Mao XZ, Feng JX, Qin YM, Zhang L, Cheng J, Wei LP, Wang ZY, Zhu YX (2006) Transcriptome profiling, molecular biological, and physiological studies reveal a major role for ethylene in cotton fiber cell elongation. Plant Cell 18(3):651–664. https://doi.org/10.1105/tpc.105.040303
Song GL, Cui RX, Wang KB, Guo LP, Zhang XD (1998) A rapid improved CTAB method for extraction of cotton genomic DNA. Acta Gossypii Sinica 10:273–275
Sparks JA, Kwon T, Renna L, Liao F, Brandizzi F, Blancaflor EB (2016) HLB1 is a tetratricopeptide repeat domain-containing protein that operates at the intersection of the exocytic and endocytic pathways at the TGN/EE in Arabidopsis. Plant Cell 28(3):746–769. https://doi.org/10.1105/tpc.15.00794
Sun W, Gao Z, Wang J, Huang Y, Chen Y, Li J, Lv M, Wang J, Luo M, Zuo K (2019) Cotton fiber elongation requires the transcription factor GhMYB212 to regulate sucrose transportation into expanding fibers. New Phytol 222(2):864–881. https://doi.org/10.1111/nph.15620
Sun X, Gong SY, Nie XY, Li Y, Li W, Huang GQ, Li XB (2015) A R2R3-MYB transcription factor that is specifically expressed in cotton (Gossypium hirsutum) fibers affects secondary cell wall biosynthesis and deposition in transgenic Arabidopsis. Physiol Plant 154(3):420–432. https://doi.org/10.1111/ppl.12317
Tian Y, Du J, Wu H, Guan X, Chen W, Hu Y, Fang L, Ding L, Li M, Yang D, Yang Q, Zhang T (2020) The transcription factor MML4_D12 regulates fiber development through interplay with the WD40-repeat protein WDR in cotton. J Exp Bot 71(12):3499–3511. https://doi.org/10.1093/jxb/eraa104
Tian Y, Zhang T (2021) MIXTAs and phytohormones orchestrate cotton fiber development. Curr Opin Plant Biol 59:101975. https://doi.org/10.1016/j.pbi.2020.10.007
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93(1):77–78. https://doi.org/10.1093/jhered/93.1.77
Wang F, Zhang J, Chen Y, Zhang C, Gong J, Song Z, Zhou J, Wang J, Zhao C, Jiao M, Liu A, Du Z, Yuan Y, Fan S, Zhang J (2020a) Identification of candidate genes for key fibre-related QTLs and derivation of favourable alleles in Gossypium hirsutum recombinant inbred lines with G. barbadense introgressions. Plant Biotechnol J 18(3):707–720. https://doi.org/10.1111/pbi.13237
Wang J, Sun W, Kong X, Zhao C, Li J, Chen Y, Gao Z, Zuo K (2020b) The peptidyl-prolyl isomerases FKBP15-1 and FKBP15-2 negatively affect lateral root development by repressing the vacuolar invertase VIN2 in Arabidopsis. Planta 252(4):52. https://doi.org/10.1007/s00425-020-03459-2
Wang L, Li XR, Lian H, Ni DA, He YK, Chen XY, Ruan YL (2010) Evidence that high activity of vacuolar invertase is required for cotton fiber and Arabidopsis root elongation through osmotic dependent and independent pathways, respectively. Plant Physiol 154(2):744–756. https://doi.org/10.1104/pp.110.162487
Wang M, Tu L, Yuan D, Zhu SC, Li J, Liu F, Pei L, Wang P, Zhao G, Ye Z, Huang H, Yan F, Ma Y, Zhang L, Liu M, You J, Yang Y, Liu Z, Huang F, Li B, Qiu P, Zhang Q, Zhu L, Jin S, Yang X, Min L, Li G, Chen LL, Zheng H, Lindsey K, Lin Z, Udall JA, Zhang X (2019a) Reference genome sequences of two cultivated allotetraploid cottons, Gossypium hirsutum and Gossypium barbadense. Nat Genet 51(2):224–229. https://doi.org/10.1038/s41588-018-0282-x
Wang N, Ma Q, Wu M, Pei W, Song J, Jia B, Liu G, Sun H, Zang X, Yu S, Zhang J, Yu J (2021) Genetic variation in MYB5_A12 is associated with fibre initiation and elongation in tetraploid cotton. Plant Biotechnol J 19(10):1892–1894. https://doi.org/10.1111/pbi.13662
Wang P, Zhang TZ (2012) Genetic dissection of photosynthetic pigment content in cotton interspecific chromosome segment introgression lines. Acta Agron Sin 38:947–953. https://doi.org/10.3724/SP.J.1006.2012.00947
Wang S, Wang JW, Yu N, Li CH, Luo B, Gou JY, Wang LJ, Chen XY (2004) Control of plant trichome development by a cotton fiber MYB gene. Plant Cell 16(9):2323–2334. https://doi.org/10.1105/tpc.104.024844
Wang Z, Yang Z, Li F (2019b) Updates on molecular mechanisms in the development of branched trichome in Arabidopsis and nonbranched in cotton. Plant Biotechnol J 17(9):1706–1722. https://doi.org/10.1111/pbi.13167
Wu M, Li L, Liu G, Li X, Pei W, Li X, Zhang J, Yu S, Yu J (2019) Differentially expressed genes between two groups of backcross inbred lines differing in fiber length developed from Upland × Pima cotton. Mol Biol Rep 46(1):1199–1212. https://doi.org/10.1007/s11033-019-04589-x
Xiao G, Zhao P, Zhang Y (2019) A pivotal role of hormones in regulating cotton fiber development. Front Plant Sci 10:87. https://doi.org/10.3389/fpls.2019.00087
Xiao YH, Li DM, Yin MH, Li XB, Zhang M, Wang YJ, Dong J, Zhao J, Luo M, Luo XY, Hou L, Hu L, Pei Y (2010) Gibberellin 20-oxidase promotes initiation and elongation of cotton fibers by regulating gibberellin synthesis. J Plant Physiol 167(10):829–837. https://doi.org/10.1016/j.jplph.2010.01.003
Xu P, Gao J, Cao Z, Chee PW, Guo Q, Xu Z, Paterson AH, Zhang X, Shen X (2017) Fine mapping and candidate gene analysis of qFL-chr1, a fiber length QTL in cotton. Theor Appl Genet 130(6):1309–1319. https://doi.org/10.1007/s00122-017-2890-8
Yang Z, Ge X, Yang Z, Qin W, Sun G, Wang Z, Li Z, Liu J, Wu J, Wang Y, Lu L, Wang P, Mo H, Zhang X, Li F (2019) Extensive intraspecific gene order and gene structural variations in upland cotton cultivars. Nat Commun 10(1):2989. https://doi.org/10.1038/s41467-019-10820-x
Yang Z, Li J (2009) Developing chromosome segment substitution lines (CSSLs) in cotton (Gossypium) using advanced backcross and MAS. Mol Plant Breed 7(2):233–241
Yang Z, Qanmber G, Wang Z, Yang Z, Li F (2020) Gossypium genomics: trends, scope, and utilization for cotton improvement. Trends Plant Sci 25(5):488–500. https://doi.org/10.1016/j.tplants.2019.12.011
Ye Z, Qiao L, Luo X, Chen X, Zhang X, Tu L (2021) Genome-wide identification of cotton GRAM family proteins reveals that GRAM31 regulates fiber length. J Exp Bot 72(7):2477–2490. https://doi.org/10.1093/jxb/eraa597
Zhang M, Wang C, Lin Q, Liu A, Wang T, Feng X, Liu J, Han H, Ma Y, Bonea D, Zhao R, Hua X (2015) A tetratricopeptide repeat domain-containing protein SSR1 located in mitochondria is involved in root development and auxin polar transport in Arabidopsis. Plant J 83(4):582–599. https://doi.org/10.1111/tpj.12911
Zhang M, Zheng X, Song S, Zeng Q, Hou L, Li D, Zhao J, Wei Y, Li X, Luo M, Xiao Y, Luo X, Zhang J, Xiang C, Pei Y (2011) Spatiotemporal manipulation of auxin biosynthesis in cotton ovule epidermal cells enhances fiber yield and quality. Nat Biotechnol 29(5):453–458. https://doi.org/10.1038/nbt.1843
Zhang Z, Ge Q, Liu A, Li J, Gong J, Shang H, Shi Y, Chen T, Wang Y, Palanga KK, Muhammad J, Lu Q, Deng X, Tan Y, Liu R, Zou X, Rashid H, Iqbal MS, Gong W, Yuan Y (2017) Construction of a high-density genetic map and its application to QTL identification for fiber strength in upland cotton. Crop Sci 57:774–788. https://doi.org/10.2135/cropsci2016.06.0544
Zhang Z, Li J, Jamshed M, Shi Y, Liu A, Gong J, Wang S, Zhang J, Sun F, Jia F, Ge Q, Fan L, Zhang Z, Pan J, Fan S, Wang Y, Lu Q, Liu R, Deng X, Zou X, Jiang X, Liu P, Li P, Iqbal MS, Zhang C, Zou J, Chen H, Tian Q, Jia X, Wang B, Ai N, Feng G, Wang Y, Hong M, Li S, Lian W, Wu B, Hua J, Zhang C, Huang J, Xu A, Shang H, Gong W, Yuan Y (2020) Genome-wide quantitative trait loci reveal the genetic basis of cotton fibre quality and yield-related traits in a Gossypium hirsutum recombinant inbred line population. Plant Biotechnol J 18(1):239–253. https://doi.org/10.1111/pbi.13191
Zhang ZS, Xiao YH, Luo M, Li XB, Luo XY, Hou L, Li DM, Pei Y (2005) Construction of a genetic linkage map and QTL analysis of fiber-related traits in upland cotton (Gossypium hirsutum L.). Euphytica 144:91–99. https://doi.org/10.1007/s10681-005-4629-x
Zhang J, Wu YT, Guo WZ, Zhang TZ (2000) Fast screening of microsatel lite markers in cotton with PAGE/silver staining. Acta Gossypii Sinica 12(5):267–269
Zhao N, Wang W, Grover CE, Jiang K, Pan Z, Guo B, Zhu J, Su Y, Wang M, Nie H, Xiao L, Guo A, Yang J, Cheng C, Ning X, Li B, Xu H, Adjibolosoo D, Aierxi A, Li P, Geng J, Wendel JF, Kong J, Hua J (2022) Genomic and GWAS analyses demonstrate phylogenomic relationships of Gossypium barbadense in China and selection for fibre length, lint percentage and Fusarium wilt resistance. Plant Biotechnol J 20(4):691–710. https://doi.org/10.1111/pbi.13747
Zhao T, Xu X, Wang M, Li C, Li C, Zhao R, Zhu S, He Q, Chen J (2019) Identification and profiling of upland cotton microRNAs at fiber initiation stage under exogenous IAA application. BMC Genomics 20(1):421. https://doi.org/10.1186/s12864-019-5760-8
Funding
This study was funded by the National Natural Science Foundation of China (32272188, 32070560), the Natural Science Foundation of Henan Province (202300410549), China Agriculture Research System of MOF and MARA, Hainan Yazhou Bay Seed Lab (B21HJ0210), the National Agricultural Science and Technology Innovation Project for CAAS (CAAS-ASTIP-2016-ICR), the Xinjiang Production and Construction Corps Innovation Program for Science and Technology Talents (2020CB005), and the National Key R&D Program for Crop Breeding (2016YFD0100306).
Author information
Authors and Affiliations
Contributions
YY, QL, and YS conceived and designed the experiments. XX, PL, ZL, WG, AL, QG, XD, YP, and SL carried out the assessment of lint percentage. XX, RL, and JG conducted the experiment. XX, QC, HZ, RP, YP, HS, and JP revised the language and manuscript. XX wrote the manuscript. All authors have read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Communicated by David D. Fang.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
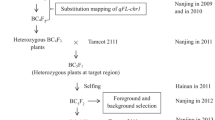
Figure S1
Program for developing the mapping population. (TIF 463 kb)
Figure S2
Frequency distribution for fiber length, fiber uniformity, fiber micronaire, fiber strength, bell weight, and lint of the primary mapping population BC6F2 (TIF 2445 kb)
Figure S3
Frequency distributions for fiber length, fiber uniformity, fiber strength, and fiber micronaire of the primary mapping population BC6F2:3. (a) Frequency distribution of the primary BC6F2:3 population in Anyang, Henan in 2017. (b) Frequency distribution of the primary BC6F2:3 population in Korla, Xinjiang, in 2017 (TIF 1813 kb)
Figure S4
Frequency distribution for fiber length, fiber uniformity, fiber strength, fiber micronaire, bell weight, and lint of the fine-mapped BC7F2 population (TIF 2375 kb)
Figure S5
Frequency distribution for fiber length, fiber uniformity, fiber strength, fiber micronaire, bell weight, and lint of the fine-mapped BC7F2:3 population (TIF 2407 kb)
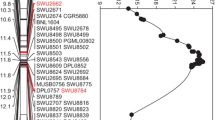
Figure S6
Chromosomal locations of QTLs of qFL-A12-5 in the primary mapping population BC6F2. NOTE: The genetic map on the left of the chromosomes indicates the genetic distance (cM) (TIF 134 kb)
Figure S7
The ~2 kb genome sequences in the upstream region of GhTPR in Hai 1, MBI774, CCRI45, and TM-1 (TIF 613 kb)
Figure S8
The Cis-acting regulatory elements (CARE) analyses in the GhTPR promoter region in Hai1 (TIF 172 kb)
Figure S9
Epidermal hairs on the stem of transgenic Arabidopsis plants overexpressing GhTPR under the Cauliflower mosaic virus (CaMV) 35S promoter. (a) Comparison of the epidermal hairs on the stem between the transgenic lines and the wild type. (b) Analysis of the number of epidermal hairs on the stem between the transgenic lines and the wild type. (c) Expression level of the GhTPR gene in the stems of the transgenic lines (TIF 7942 kb)
Table S1
Phenotypic analysis of fiber quality and yield traits in the primary populations of BC6F2 and BC6F2:3 (DOCX 21 kb)
Table S2
Correlation coefficients corresponding to fiber length (FL) traits in the mapping populations (DOCX 15 kb)
Table S3
ANOVA analysis for fiber length (FL) traits in the mapping populations (DOCX 16 kb)
Table S4
qRT-PCR primers for candidate genes in the qFL-A12-5 interval were developed from the G. hirsutum genome sequence (DOCX 16 kb)
Table S5
Clone primers for candidate genes in the qFL-A12-5 interval were developed from the G. hirsutum genome sequence (DOCX 16 kb)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xiao, X., Liu, R., Gong, J. et al. Fine mapping and candidate gene analysis of qFL-A12-5: a fiber length-related QTL introgressed from Gossypium barbadense into Gossypium hirsutum. Theor Appl Genet 136, 48 (2023). https://doi.org/10.1007/s00122-023-04247-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00122-023-04247-8