Abstract
Key message
In this study, we fine-mapped a clubroot resistance gene CRA3.7 in Chinese cabbage and developed its closely linked marker syau-InDel3008 for marker-assisted selection in CR cultivars breeding.
Abstract
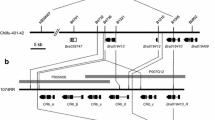
Chinese cabbage is an important leafy vegetable rich in many nutrients widely grown in China. Clubroot disease caused by an obligate biotrophic pathogen Plasmodiophora brassicae was rapidly spread and challenged to Chinese cabbage production. A clubroot resistance (CR) gene, CRA3.7, was mapped on chromosome A03 of Brassica rapa. A Chinese cabbage line ‘CR510’, which harbor homozygous resistance locus CRA3.7 was selected from a BC4F3 family. ‘CR510’ was crossed with a clubroot susceptible Chinese cabbage inbred line ‘59-1’. Total 51 recombinant plants were identified from an F2 population including 3000 individuals. These recombinants were selfed and the clubroot resistance of F2/3 families was evaluated. Finally, a clubroot resistance gene CRA3.7 was fine-mapped to an interval of approximately 386 kb between marker syau-InDel3024 and syau-InDel3008. According to the reference genome, total 54 genes including five encoding the TIR-NBS-LRR proteins was annotated in the fine-mapped region. Further, nine candidate’s gene expression in parental lines at 7, 14 and 21 days after inoculation of P. brassicae were evaluated. Bra019376, Bra019401, Bra019403 and Bra019410 are highly expressed in ‘CR510’ than ‘59-1’. Gene sequence of Bra019410 from ‘CR510’ was cloned and identified different from CRa. Therefore, Bra019376, Bra019401, Bra019403 and Bra019410 are the most likely candidates for CRA3.7. Our research provides a valuable germplasm resource against P. brassicae Pb3 and CRA3.7 closely linked marker for marker-assisted selection in CR cultivars breeding.


Similar content being viewed by others
Data availability
The datasets generated and analyzed during this study are available on reasonable requests from the corresponding authors. The raw data of whole genome re-sequencing have been deposited in the NCBI Sequence Read Archive (SRA) repository under the accession numbers SAMN28869001 and SAMN28869002.
References
Aruga D, Ueno H, Matsumura H, Matsumoto E, Hayashida N (2013) Distribution of CRa in clubroot resistance (CR) cultivars of Chinese cabbage. Plant Biotechnol. https://doi.org/10.5511/plantbiotechnology.13.0110a
Chen J, Jing J, Zhan Z, Zhang T, Zhang C, Piao Z (2013) Identification of novel QTLs for isolate-specific partial resistance to Plasmodiophora brassicae in Brassica rapa. PLoS One 8:e85307. https://doi.org/10.1371/journal.pone.0085307
Diederichsen E, Frauen M, Linders EGA, Hatakeyama K, Hirai M (2009) Status and perspectives of clubroot resistance breeding in crucifer crops. J Plant Growth Regul 28:265–281
Dixon GR (2009) The occurrence and economic impact of Plasmodiophora brassicae and clubroot disease. J Plant Growth Regul 28:194–202
Dolatabadian A, Cornelsen J, Huang S, Zou Z, Fernando WGD (2021) Sustainability on the farm: breeding for resistance and management of major canola diseases in Canada contributing towards an IPM approach. Can J Plant Pathol 44(2):157–190
Drury SC, Gossen BD, McDonald MR (2021) Clubroot resistance in canola and brassica vegetable cultivars in Ontario. Canada. Can J Plant Sci 101(5):730–740
Hatakeyama K, Suwabe K, Tomita RN, Kato T, Nunome T, Fukuoka H, Matsumoto S (2013) Identification and characterization of Crr1a, a gene for resistance to clubroot disease (Plasmodiophora brassicae Woronin) in Brassica rapa L. PLoS One 8:e54745. https://doi.org/10.1371/journal.pone.0054745
Hatakeyama K, Niwa T, Kato T, Ohara T, Kakizaki T, Matsumoto S (2017) The tandem repeated organization of NB-LRR genes in the clubroot resistant CRb, locus in Brassica rapa L. Mol Genet Genomics 292:397–405. https://doi.org/10.1007/s00438-016-1281-1
Hirani AH, Gao F, Liu J, Fu G, Wu C, McVetty PBE, Duncan R, Li G (2018) Combinations of independent dominant loci conferring clubroot resistance in all four turnip accessions (Brassica rapa) from the European clubroot differential set. Front Plant Sci 9:1628. https://doi.org/10.3389/fpls.2018.01628
Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK (2012) VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 22(3):568–76
Kuginuki Y, Yoshigawa H, Hirai M (1999) Variation in virulence of Plasmodiophora brassicae in Japan tested with clubroot resistant cultivars of Chinese cabbage. Eur J Plant Pathol 105:327–332. https://doi.org/10.1023/A:1008705413127
Larkan NJ, Ma L, Haddadi P, Buchwaldt M, Parkin IAP, Djavaheri M, Borhan MH (2020) The Brassica napus wall-associated kinase-like (WAKL) gene Rlm9 provides race-specific blackleg resistance. Plant J. https://doi.org/10.1111/tpj.14966
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–60
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup (2009) The sequence alignment/Map format and SAMtools. Bioinformatics 25:2078–2079
Lv H, Fang Z, Yang L, Zhang Y, Wang Y (2020) An update on the arsenal: mining resistance genes for disease management of Brassica crops in the genomic era. Hortic Res 7:34
Ma Y, Choi SR, Wang Y, Chhapekar SS, Zhang X, Wang Y, Zhang X, Zhu M, Liu D, Zuo Z, Yan X, Gan C, Zhao D, Liang Y, Pang W, Lim YP (2022) Starch content changes and metabolism-related gene regulation of Chinese cabbage synergistically induced by plasmodiophora brassicae infection. Hortic Res. https://doi.org/10.1093/hr/uhab071
Ning J, Pang W, Chen B, Zhan Z, Li Q, Li M, Zhang C, Piao Z (2015) Validation of a quantitative trait locus QS_B3.1 for clubroot resistance in near-isogenic lines of Chinese cabbage. Mol Plant Breed 13(12):2782–2787
Van Ooijen JW, Voorrips RE (2001) Join map version 3.0: Software for the calculation of genetic linkage maps. In: Wageningen: plant research international
Pang W, Liang S, Li X, Li P, Yu S, Lim YP et al (2014) Genetic detection of clubroot resistance loci in a new population of Brassica rapa. Hortic Environ Biotechnol 55:540–547. https://doi.org/10.1007/s13580-014-0079-5
Pang W, Fu P, Li X, Zhan Z, Yu S, Piao Z (2018) Identification and mapping of the clubroot resistance gene CRd in Chinese cabbage (Brassica rapa ssp. pekinensis). Front Plant Sci 9:653
Pang W, Liang Y, Zhan Z, Li X, Piao Z (2020) Development of a sinitic clubroot differential set for the pathotype classification of Plasmodiophora brassicae. Front Plant Sci 11:568771
Piao ZY, Deng YQ, Choi SR, Park YJ, Lim YP (2004) SCAR and CAPS mapping of CRb, a gene conferring resistance to Plasmodiophora brassicae in Chinese cabbage (Brassica rapa ssp. pekinensis). Theor Appl Genet 108:1458–1465
Rocherieux J, Glory P, Giboulot A, Boury S, Barbeyron G, Thomas G, Manzanares-Dauleux MJ (2004) Isolate-specific and broad-spectrum QTLs are involved in the control of clubroot in Brassica oleracea. Theor Appl Genet 108:1555–1563
Sakamoto K, Saito A, Hayashida N, Taguchi G, Matsumoto E (2008) Mapping of isolate-specific QTLs for clubroot resistance in Chinese cabbage (Brassica rapa L. ssp. pekinensis). Theor Appl Genet 117:759–767
Song X, Hu J, Wu T, Yang Q, Feng X, Lin H, Feng S, Cui C, Yu Y, Zhou R, Gong K, Tong Yu, Pei Q, Li N (2021) Comparative analysis of long noncoding RNAs in angiosperms and characterization of long noncoding RNAs in response to heat stress in Chinese cabbage. Hortic Res 8:48
Stam P (1993) Construction of integrated genetic linkage maps by means of a new computer package: joinmap. Plant J 3:739–744
Strelkov SE, Hwang S, Manolii VP, Cao T, Feindel D (2016) Emergence of new virulence phenotypes of Plasmodiophora brassicae on canola (Brassica napus) in Alberta. Can Eur J Plant Pathol 145:517–529. https://doi.org/10.1007/s10658-016-0888-8
Suwabe K, Tsukazaki H, Iketani H, Hatakeyama K, Fujimura M, Nunome T, Fukuoka H, Matsumoto S, Hirai M (2003) Identification of two loci for resistance to clubroot (Plasmodiophora brassicae Woronin) in Brassica rapa L. Theor Appl Genet 107:997–1002
Suwabe K, Tsukazaki H, Iketani H, Hatakeyama K, Kondo M, Fujimura M, Nunome T, Fukuoka H, Hirai M, Matsumoto S (2006) Simple sequence repeat-based comparative genomics between Brassica rapa and Arabidopsis thaliana: the genetic origin of clubroot resistance. Genetics 173:309–319
Ueno H, Matsumoto E, Aruga D, Kitagawa S, Matsumura H, Hayashida N (2012) Molecular characterization of the CRa gene conferring clubroot resistance in Brassica rapa. Plant Mol Biol 80:621–629
Wang X, Wang H, Wang J, Sun R, Wu J, Liu S, Bai Y, Mun J-H, Bancroft I, Cheng F et al (2011) The genome of the mesopolyploid crop species Brassica rapa. Nat Genet 43:1035–1039
Wang Y, Xiang X, Huang F, Yu W, Zhou X, Li B, Zhang Y, Chen P, Zhang C (2022) Fine mapping of clubroot resistance loci CRA8.1 and candidate gene analysis in Chinese cabbage (Brassica rapa L.). Front Plant Sci 13:898108. https://doi.org/10.3389/fpls.2022.898108
Werner S, Diederichsen E, Frauen M, Schondelmaier J, Jung C (2008) Genetic mapping of clubroot resistance genes in oilseed rape. Theor Appl Genet 116:363–372
Williams PH (1966) A system for the determination of races of Plasmodiophora brassicae that infect cabbage and rutabaga. Phytopathology 56:624–626
Yang Z, Jiang Y, Gong J, Li Q, Dun B, Liu D, Yin F, Yuan L, Zhou X, Wang H, Wang J, Zhan Z, Shah N, Nwafor CC, Zhou Y, Chen P, Zhu L, Li S, Wang B, Xiang J, Zhou Y, Li Z, Pia Z, Yang Q, Zhang C (2022) R gene triplication confers European fodder turnip with improved clubroot resistance. Plant Biotechnol J. https://doi.org/10.1111/pbi.13827
Acknowledgment
The authors thank Prof. Yue Liang for the construction suggestions on the writing of this paper, as well as the laboratory members for assistance with the plant materials growing and surveys of clubroot resistance tests.
Funding
This study was supported by grants from the National Natural Science Foundation of China, Project No. 32272720; the Liaoning Natural Science Foundation (2021-MS-229), the China Postdoctoral Science Foundation (2016M600214).
Author information
Authors and Affiliations
Contributions
XZ, WXP and YW carried out experiments, generated data. WXP analyzed the data and wrote the original manuscript. YM and ZZ participated in data analysis. WXP and ZYP conceived the study, participated in its coordination. All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Communicated by Jacqueline Batley.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Pang, W., Zhang, X., Ma, Y. et al. Fine mapping and candidate gene analysis of CRA3.7 conferring clubroot resistance in Brassica rapa. Theor Appl Genet 135, 4541–4548 (2022). https://doi.org/10.1007/s00122-022-04237-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-022-04237-2