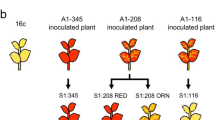
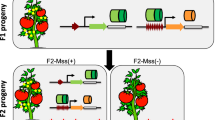
Abstract
Key message
Crosses of parents that differ in their DNA methylation states leads to progressive demethylation in the F1 hybrids.
Abstract
In plant breeding research, hybrid vigor in F1 hybrids is known to be a very important phenomenon. Hybrid vigor, or heterosis, refers to the fact that F1 hybrids from crosses with a certain combination of parents have traits that are superior to those of the parents. In addition, DNA methylation is an important factor that affects gene expression in plant genomes and contributes to hybrid vigor. We introduced the 35S promoter sequence into the cucumber mosaic virus (CMV)-based vector and inoculated the GFP-expressing transgenic Nicotiana benthamiana line 16c with the recombinant virus specifically to induce DNA methylation on the 35S promoter. For plants that had transcriptional gene silencing (TGS) of GFP established by methylation of the 35S promoter (35S-TGS), TGS was fully maintained in their later self-pollinated generations. When the 35S-TGS plants were crossed with 16c, which does not contain DNA methylation in the 35S promoter, the F1 hybrids unexpectedly became progressively DNA demethylated as the plants grew. We hypothesis that in F1 hybrids that are produced by a cross between parents with extremely different gene methylation states, the methylation state of the genes in question may shift more and more to hypomethylation as the plants grow. This progressive demethylation phenomenon observed in this study may be important in plant breeding to reactivate the genes which were silenced by DNA methylation.





Similar content being viewed by others
Availability of data and materials
All data generated and analyzed during this study are included in this published article.
References
Baulcombe D (2004) RNA silencing in plants. Nature 431:356–363
Benfey PN, Chua NH (1990) The cauliflower mosaic virus 35S promoter: combinatorial regulation of transcription in plants. Science 250:959–966
Borges F, Martienssen RA (2013) Establishing epigenetic variation during genome reprogramming. RNA Biol 10:490–494
Calarco JP, Borges F, Donoghue M, Ex FV, Jullien PE, Lopes T, Gardner R, Berger F, Feijó JA, Becker JD, Martienssen RA (2012) Reprogramming of DNA methylation in pollen guides epigenetic inheritance via small RNA. Cell 151:194–205
Cao X, Aufsatz W, Zilberman D, Mette MF, Huang MS, Matzke M, Jacobsen SE (2003) Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr Biol 13:2212–2217
Dalakouras A, Vlachostergios D (2021) Epigenetic approaches to crop breeding: current status and perspectives. J Exp Bot 28(72):5356–5371
Fujimoto R, Taylor JM, Shirasawa S, Peacock WJ, Dennis ES (2012) Heterosis of Arabidopsis hybrids between C24 and Col is associated with increased photosynthesis capacity. Proc Natl Acad Sci USA 109:7109–7114
Fujimoto R, Uezono K, Ishikura S, Osabe K, Peacock WJ, Dennis ES (2018) Recent research on the mechanism of heterosis is important for crop and vegetable breeding systems. Breed Sci 68:145–158
Gehring M (2019) Epigenetic dynamics during flowering plant reproduction: evidence for reprogramming? New Phytol 224:91–96
Greaves IK, Bayon RG, Wang L, Zhu A, Liu PC, Groszmann M, Peacock WJ, Dennis ES (2015) Epigenetic changes in hybrids. Plant Physiol 168:1197–1205
Greaves IK, Eichten SR, Groszmann M, Wang A, Ying H, Peacock WJ, Dennis ES (2016) Twenty-four-nucleotide siRNAs produce heritable trans-chromosomal methylation in F1 Arabidopsis hybrids. Proc Natl Acad Sci USA 113:6895–6902
Greaves IK, Groszmann M, Wang A, Peacock WJ, Dennis ES (2014) Inheritance of trans chromosomal methylation patterns from Arabidopsis F1 hybrids. Proc Natl Acad Sci USA 111:2017–2022
Greaves IK, Groszmann M, Ying H, Taylor JM, James Peacock WJ, Dennis ES (2012) Trans chromosomal methylation in Arabidopsis hybrids. Proc Natl Acad Sci USA 109:3570–3575
Groszmann M, Greaves IK, Albert N, Fujimoto R, Helliwell CA, Dennis ES, Peacock WJ (2011a) Epigenetics in plants-vernalisation and hybrid vigour. Biochim Biophys Acta 1809:427–437
Groszmann M, Greaves IK, Albertyn ZI, Scofield GN, Peacock WJ, Dennis ES (2011b) Changes in 24-nt siRNA levels in Arabidopsis hybrids suggest an epigenetic contribution to hybrid vigor. Proc Natl Acad Sci USA 108:2617–2622
Groszmann M, Greaves IK, Fujimoto R, Peacock WJ, Dennis ES (2013) The role of epigenetics in hybrid vigour. Trends Genet 12:684–690
Ibarra CA, Feng X, Schoft VK, Hsieh TF, Uzawa R, Rodrigues JA, Zemach A, Chumak N, Machlicova A, Nishimura T, Rojas D, Fischer RL, Tamaru H, Zilberman D (2012) Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science 337:1360–1364
Ingouff M, Selles B, Michaud C, Vu TM, Berger F, Schorn AJ, Autran D, Durme MV, Nowack MK, Martienssen RA, Grimanelli D (2017) Live-cell analysis of DNA methylation during sexual reproduction in Arabidopsis reveals context and sex-specific dynamics controlled by noncanonical RdDM. Genes Dev 31:72–83
Jullien PE, Kinoshita T, Ohad N, Berger F (2006) Maintenance of DNA methylation during the Arabidopsis life cycle is essential for parental imprinting. Plant Cell 18:1360–1372
Jullien PE, Mosquna A, Ingouff M, Sakata T, Ohad N, Berger F (2008) Retinoblastoma and its binding partner MSI1 control imprinting in Arabidopsis. PLoS Biol 6:e194
Jullien PE, Susaki D, Yelagandula R, Higashiyama T, Berger F (2012) DNA methylation dynamics during sexual reproduction in Arabidopsis thaliana. Curr Biol 22:1825–1830
Kawakatsu T, Nery JR, Castanon R, Ecker JR (2017) Dynamic DNA methylation reconfiguration during seed development and germination. Genome Biol 18:171
Kawanabe T, Ishikura S, Miyaji N, Sasaki T, Wu LM, Itabashi E, Takada S, Shimizu M, Takasaki-Yasuda T, Osabe K, Peacock WJ, Dennis ES, Fujimoto R (2016) Role of DNA methylation in hybrid vigor in Arabidopsis thaliana. Proc Natl Acad Sci USA 113:6704–6711
Kawashima T, Berger F (2014) Epigenetic reprogramming in plant sexual reproduction. Nat Rev Genet 15:613–624
Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Luo S, Preuss D (2003) Strand-biased DNA methylation associated with centromeric regions in Arabidopsis. Proc Natl Acad Sci USA 100:11133–11138
Matsunaga W, Shimura H, Shirakawa S, Isoda R, Inukai T, Matsumura T, Masuta C (2019) Transcriptional silencing of 35S driven-transgene is differentially determined depending on promoter methylation heterogeneity at specific cytosines in both plus- and minus-sense strands. BMC Plant Biol 19:24
Matzke MA, Kanno T, Matzke AJ (2015) RNA-Directed DNA Methylation: the evolution of a complex epigenetic pathway in flowering plants. Annu Rev Plant Biol 66:243–267
Otagaki S, Arai M, Takahashi A, Goto K, Hong JS, Masuta C (2006) Rapid induction of transcriptional and post-transcriptional gene silencing using a novel Cucumber mosaic virus vector. Plant Biotechnol 23:259–265
Shen H, He H, Li J, Chen W, Wang X, Guo L, Peng Z, He G, Zhong S, Qi Y, Terzaghi W, Deng XW (2012) Genome-wide analysis of DNA methylation and gene expression changes in two Arabidopsis ecotypes and their reciprocal hybrids. Plant Cell 24:875–892
Wang XL, Song SH, Wu YS, Li YL, Chen TT, Huang ZY, Liu S, Dunwell TL, Pfeifer GP, Dunwell JM, Wamaedeesa R, Ullah I, Wang Y, Hu SN (2015) Genome-wide mapping of 5-hydroxymethylcytosine in three rice cultivars reveals its preferential localization in transcriptionally silent transposable element genes. J Exp Bot 66:6651–6663
Zhang Q, Wang D, Lang Z, He L, Yang L, Zeng L, Li Y, Zhao C, Huang H, Zhang H, Zhang H, Zhu JK (2016) Methylation interactions in Arabidopsis hybrids require RNA-directed DNA methylation and are influenced by genetic variation. Proc Natl Acad Sci USA 113:4248–4256
Zhang Y, Harris CJ, Liu Q, Liu W, Ausin I, Long Y, Xiao L, Feng L, Chen X, Xie Y, Chen X, Zhan L, Feng S, Li JJ, Wang H, Zhai J, Jacobsen SE (2018) Large-scale comparative epigenomics reveals hierarchical regulation of non-CG methylation in Arabidopsis. Proc Natl Acad Sci USA 115:1069–1074
Zhong X, Du J, Hale CJ, Gallego-Bartolome J, Feng S, Vashisht AA, Chory J, Wohlschlegel JA, Patel DJ, Jacobsen SE (2014) Molecular mechanism of action of plant DRM de novo DNA methyltransferases. Cell 157:1050–1060
Zhou S, Xing M, Zhao Z, Gu Y, Xiao Y, Liu Q, Xu H (2021) DNA methylation modification in heterosis initiation through analyzing rice hybrid contemporary seeds. Crop J. https://doi.org/10.1016/j.cj.2020.12.003
Funding
This work was supported by the promotion services of the New Energy and Industrial Technology Development Organization (NEDO) (project code: P16009).
Author information
Authors and Affiliations
Contributions
CM and TI designed and coordinated the study. WM performed all analyses. All authors contributed to the analysis of results, manuscript revisions and approved the final publication.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interests.
Additional information
Communicated by Kan Wang.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Matsunaga, W., Inukai, T. & Masuta, C. Progressive DNA demethylation in epigenetic hybrids between parental plants with and without methylation of the transgene promoter. Theor Appl Genet 135, 883–893 (2022). https://doi.org/10.1007/s00122-021-04004-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-021-04004-9