Abstract
Key message
Using phenotype data of three spring wheat populations evaluated at 6–15 environments under two management systems, we found moderate to very high prediction accuracies across seven traits. The phenotype data collected under an organic management system effectively predicted the performance of lines in the conventional management and vice versa.
Abstract
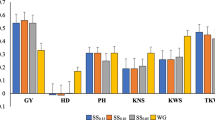
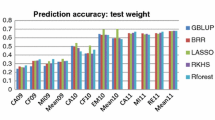
There is growing interest in developing wheat cultivars specifically for organic agriculture, but we are not aware of the effect of organic management on the predictive ability of genomic selection (GS). Here, we evaluated within populations prediction accuracies of four GS models, four combinations of training and testing sets, three reaction norm models, and three random cross-validations (CV) schemes in three populations phenotyped under organic and conventional management systems. Our study was based on a total of 578 recombinant inbred lines and varieties from three spring wheat populations, which were evaluated for seven traits at 3–9 conventionally and 3–6 organically managed field environments and genotyped either with the wheat 90 K SNP array or DArTseq. We predicted the management systems (CV0M) or environments (CV0), a subset of lines that have been evaluated in either management (CV2M) or some environments (CV2), and the performance of newly developed lines in either management (CV1M) or environments (CV1). The average prediction accuracies of the model that incorporated genotype × environment interactions with CV0 and CV2 schemes varied from 0.69 to 0.97. In the CV1 and CV1M schemes, prediction accuracies ranged from − 0.12 to 0.77 depending on the reaction norm models, the traits, and populations. In most cases, grain protein showed the highest prediction accuracies. The phenotype data collected under the organic management effectively predicted the performance of lines under conventional management and vice versa. This is the first comprehensive GS study that investigated the effect of the organic management system in wheat.




Similar content being viewed by others
References
An D, Su J, Liu Q, Zhu Y, Tong Y, Li J, Jing R, Li B, Li Z (2006) Mapping QTLs for nitrogen uptake in relation to the early growth of wheat (Triticum aestivum L.). Plant Soil 284:73–84
Arojju SK, Cao M, Trolove M, Barrett BA, Inch C, Eady C, Stewart A, Faville MJ (2020) Multi-trait genomic prediction improves predictive ability for dry matter yield and water-soluble carbohydrates in perennial ryegrass. Front Plant Sci 11:1197
Asif M, Yang RC, Navabi A, Iqbal M, Kamran A, Lara EP, Randhawa H, Pozniak C, Spaner D (2015) Mapping QTL, selection differentials, and the effect of Rht-B1 under organic and conventionally managed systems in the Attila × CDC Go spring wheat mapping population. Crop Sci 55:1129–1142
Baenziger PS, Salah I, Little RS, Santra DK, Regassa T, Wang MY (2011) Structuring an efficient organic wheat breeding program. Sustainability 3:1190–1205
Bassi FM, Bentley AR, Charmet G, Ortiz R, Crossa J (2016) Breeding schemes for the implementation of genomic selection in wheat (Triticum spp.). Plant Sci 242:23–36
Battenfield SD, Guzmán C, Chris Gaynor R, Singh RP, Peña RJ, Dreisigacker S, Fritz AK, Poland JA (2016) Genomic selection for processing and end-use quality traits in the CIMMYT spring bread wheat breeding program. Plant Genome 9. https://doi.org/10.3835/plantgenome2016.01.0005
Beales J, Turner A, Griffiths S, Snape JW, Laurie DA (2007) A pseudo-response regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L.). Theor Appl Genet 115:721–733
Bemister DH, Semagn K, Iqbal M, Randhawa H, Strelkov SE, Spaner DM (2019) Mapping QTL associated with stripe rust, leaf rust, and leaf spotting in a Canadian spring wheat population. Crop Sci 59:650–658
Beukert U, Thorwarth P, Zhao Y, Longin CFH, Serfling A, Ordon F, Reif JC (2020) Comparing the potential of marker-assisted selection and genomic prediction for improving rust resistance in hybrid wheat. Front Plant Sci 11:594113–594113
Beyene Y, Semagn K, Mugo S, Tarekegne A, Babu R, Meisel B, Sehabiague P, Makumbi D, Magorokosho C, Oikeh S, Gakunga J, Vargas M, Olsen M, Prasanna BM, Banziger M, Crossa J (2015) Genetic gains in grain yield through genomic selection in eight bi-parental maize populations under drought stress. Crop Sci 55:154–163
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635
Brasier K, Ward B, Smith J, Seago J, Oakes J, Balota M, Davis P, Fountain M, Brown-Guedira G, Sneller C, Thomason W, Griffey C (2020) Identification of quantitative trait loci associated with nitrogen use efficiency in winter wheat. PLoS ONE 15:e0228775
Burgueno J, de los Campos G, Weigel K, Crossa J (2012) Genomic prediction of breeding values when modeling genotype × environment interaction using pedigree and dense molecular markers. Crop Sci 52:707–719
Ceballos H, Kawuki RS, Gracen VE, Yencho GC, Hershey CH (2015) Conventional breeding, marker-assisted selection, genomic selection and inbreeding in clonally propagated crops: a case study for cassava. Theor Appl Genet 128:1647–1667
Chen F, Gao M, Zhang J, Zuo A, Shang X, Cui D (2013) Molecular characterization of vernalization and response genes in bread wheat from the Yellow and Huai Valley of China. BMC Plant Biol 13:199
Chen Z, Liu C, Wang Y, He T, Gao R, Xu H, Guo G, Li Y, Zhou L, Lu R, Huang J (2018) Expression analysis of nitrogen metabolism-related genes reveals differences in adaptation to low-nitrogen stress between two different barley cultivars at seedling stage. Int J Genom 2018:8152860
Chen H, Bemister DH, Iqbal M, Strelkov SE, Spaner DM (2020) Mapping genomic regions controlling agronomic traits in spring wheat under conventional and organic managements. Crop Sci 60:2038–2052
Clark SA, Hickey JM, van der Werf JHJ (2011) Different models of genetic variation and their effect on genomic evaluation. Genet Sel Evol 43:18
Cobb JN, Juma RU, Biswas PS, Arbelaez JD, Rutkoski J, Atlin G, Hagen T, Quinn M, Ng EH (2019) Enhancing the rate of genetic gain in public-sector plant breeding programs: lessons from the breeder’s equation. Theor Appl Genet 132:627–645
Collard BCY, Jahufer MZZ, Brouwer JB, Pang ECK (2005) An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for crop improvement: the basic concepts. Euphytica 142:169–196
Consortium RD, Fugeray-Scarbel A, Bastien C, Dupont-Nivet M, Lemarié S (2021) Why and how to switch to genomic selection: lessons from plant and animal breeding experience. Front Genet 12:1185
Crossa J, Beyene Y, Semagn K, Perez P, Hickey JM, Chen C, de los Campos G, Burgueno J, Windhausen VS, Buckler E, Jannink JL (2013) Genomic prediction in maize breeding populations with genotyping-by-sequencing. Genes Genom Genet 3:1903–1926
Crossa J, Jarquín D, Franco J, Pérez-Rodríguez P, Burgueño J, Saint-Pierre C, Vikram P, Sansaloni C, Petroli C, Akdemir D, Sneller C, Reynolds M, Tattaris M, Payne T, Guzman C, Peña RJ, Wenzl P, Singh S (2016) Genomic prediction of gene bank wheat landraces. Genes Genom Genet 6:1819–1834
Crossa J, de los Campos G, Maccaferri M, Tuberosa R, Burgueño J, Pérez-Rodríguez P (2016) Extending the marker × environment interaction model for genomic-enabled prediction and genome-wide association analysis in durum wheat. Crop Sci 56:2193–2209
Cuevas J, Crossa J, Soberanis V, Pérez-Elizalde S, Pérez-Rodríguez P, Campos GDL, Montesinos-López OA (2016) Genomic prediction of genotype × environment interaction kernel regression models. Plant Genome 9(plantgenome2016.2003):0024
Cuevas J, Crossa J, Montesinos-López OA, Burgueño J, Pérez-Rodríguez P, Delos Campos G (2017) Bayesian genomic prediction with genotype strong × strong environment interaction kernel models. Genes Genom Genet 7:41
Das RR, Vinayan MT, Patel MB, Phagna RK, Singh SB, Shahi JP, Sarma A, Barua NS, Babu R, Seetharam K, Burgueño JA, Zaidi PH (2020) Genetic gains with rapid-cycle genomic selection for combined drought and waterlogging tolerance in tropical maize (Zea mays L.). Plant Genome 13:e20035
Dawson JC, Endelman JB, Heslot N, Crossa J, Poland J, Dreisigacker S, Manès Y, Sorrells ME, Jannink J-L (2013) The use of unbalanced historical data for genomic selection in an international wheat breeding program. Field Crops Res 154:12–22
Dreisigacker S, Crossa J, Pérez-Rodríguez P, Montesinos-López OA, Rosyara U, Juliana P, Mondal S, Crespo-Herrera L, Govindan V, Singh RP, Braun HJ (2021) Implementation of genomic selection in the CIMMYT global wheat program, findings from the past 10 years. Crop Breed Genet Genom 3:e210004
Ellis MH, Spielmeyer W, Gale KR, Rebetzke GJ, Richards RA (2002) Perfect markers for the Rht-B1b and Rht-D1b dwarfing genes in wheat. Theor Appl Genet 105:1038–1042
Gelli M, Mitchell SE, Liu K, Clemente TE, Weeks DP, Zhang C, Holding DR, Dweikat IM (2016) Mapping QTLs and association of differentially expressed gene transcripts for multiple agronomic traits under different nitrogen levels in sorghum. BMC Plant Biol 16:16
Good AG, Beatty PH (2011) Fertilizing nature: a tragedy of excess in the commons. PLoS Biol 9:e1001124
Guo Y, Kong Fm Xu, Yf ZY, Liang X, Yy W, Dg An, Ss Li (2012) QTL mapping for seedling traits in wheat grown under varying concentrations of N, P and K nutrients. Theor Appl Genet 124:851–865
Haile TA, Walkowiak S, N’Diaye A, Clarke JM, Hucl PJ, Cuthbert RD, Knox RE, Pozniak CJ (2021) Genomic prediction of agronomic traits in wheat using different models and cross-validation designs. Theor Appl Genet 134:381–398
Haile JK, N’Diaye A, Clarke F, Clarke J, Knox R, Rutkoski J, Bassi FM, Pozniak CJ (2018) Genomic selection for grain yield and quality traits in durum wheat. Mol Breed 38:75
Hayes BJ, Panozzo J, Walker CK, Choy AL, Kant S, Wong D, Tibbits J, Daetwyler HD, Rochfort S, Hayden MJ, Spangenberg GC (2017) Accelerating wheat breeding for end-use quality with multi-trait genomic predictions incorporating near infrared and nuclear magnetic resonance-derived phenotypes. Theor Appl Genet 130:2505–2519
Heffner EL, Lorenz AJ, Jannink JL, Sorrells ME (2010) Plant breeding with genomic selection: gain per unit time and cost. Crop Sci 50:1681–1690
Heslot N, Akdemir D, Sorrells ME, Jannink JL (2013) Integrating environmental covariates and crop modeling into the genomic selection framework to predict genotype by environment interactions. Theor Appl Genet 127:463–480
Ibba MI, Crossa J, Montesinos-López OA, Montesinos-López A, Juliana P, Guzman C, Delorean E, Dreisigacker S, Poland J (2020) Genome-based prediction of multiple wheat quality traits in multiple years. Plant Genome 13:e20034
Jaganathan D, Bohra A, Thudi M, Varshney RK (2020) Fine mapping and gene cloning in the post-NGS era: advances and prospects. Theor Appl Genet 133:1791–1810
Jarquín D (2012) Lemes da Silva C, Gaynor RC, Poland J, Fritz A, Howard R, Battenfield S, Crossa J (2017) Increasing genomic-enabled prediction accuracy by modeling genotype × environment interactions in Kansas wheat. Plant Genome 10(plantgenome2016):0130@@@@
Jarquin D, Crossa J, Lacaze X, Cheyron P, Jl D, Lorgeou J, Fc P, Guerreiro L, Perez P, Calus M, Burgueno J, Campos G (2014) A reaction norm model for genomic selection using high-dimensional genomic and environmental data. Theor Appl Genet 127:595–607
Jarquin D, Howard R, Crossa J, Beyene Y, Gowda M, Martini JWR, Covarrubias Pazaran G, Burgueño J, Pacheco A, Grondona M, Wimmer V, Prasanna BM (2020) Genomic prediction enhanced sparse testing for multi-environment trials. Genes Genom Genet 10:2725
Juliana P, Singh RP, Braun H-J, Huerta-Espino J, Crespo-Herrera L, Govindan V, Mondal S, Poland J, Shrestha S (2020) Genomic selection for grain yield in the CIMMYT wheat breeding program—status and perspectives. Front Plant Sci 11:1418
Kirk AP, Fox SL, Entz MH (2012) Comparison of organic and conventional selection environments for spring wheat. Plant Breed 131:687–694
Krishnappa G, Savadi S, Tyagi BS, Singh SK, Mamrutha HM, Kumar S, Mishra CN, Khan H, Gangadhara K, Uday G, Singh G, Singh GP (2021) Integrated genomic selection for rapid improvement of crops. Genomics 113:1070–1086
Lado B, Vázquez D, Quincke M, Silva P, Aguilar I, Gutiérrez L (2018) Resource allocation optimization with multi-trait genomic prediction for bread wheat (Triticum aestivum L.) baking quality. Theor Appl Genet 131:2719–2731
Laidig F, Piepho H-P, Rentel D, Drobek T, Meyer U, Huesken A (2017) Breeding progress, environmental variation and correlation of winter wheat yield and quality traits in German official variety trials and on-farm during 1983–2014. Theor Appl Genet 130:223–245
Lammerts van Bueren ET, Jones SS, Tamm L, Murphy KM, Myers JR, Leifert C, Messmer MM (2011) The need to breed crop varieties suitable for organic farming, using wheat, tomato and broccoli as examples: a review. Wagen J Life Sci 58:193–205
Laperche A, Brancourt-Hulmel M, Heumez E, Gardet O, Hanocq E, Devienne-Barret FD, Gouis JL (2007) Using genotype × nitrogen interaction variables to evaluate the QTL involved in wheat tolerance to nitrogen constraints. Theor Appl Genet 115:399–415
Lian L, Jacobson A, Zhong S, Bernardo R (2014) Genomewide prediction accuracy within 969 maize biparental populations. Crop Sci 54:1514–1522
Lopez-Cruz M, Crossa J, Bonnett D, Dreisigacker S, Poland J, Jannink JL, Singh RP, Autrique E, de los Campos G (2015) Increased prediction accuracy in wheat breeding trials using a marker × environment interaction genomic selection model. Genes Genom Genet 5:569–582
Mason HE, Spaner D (2006) Competitive ability of wheat in conventional and organic management systems: a review of the literature. Can J Plant Sci 86:333–343
Mason HE, Navabi A, Frick BL, O’Donovan JT, Spaner DM (2007) The weed-competitive ability of Canada western red spring wheat cultivars grown under organic management. Crop Sci 47:1167–1176
Meuwissen THE, Hayes BJ, Goddard ME (2001) Prediction of total genetic value using genome-wide dense marker maps. Genetics 157:1819–1829
Michel S, Löschenberger F, Ametz C, Pachler B, Sparry E, Bürstmayr H (2019a) Combining grain yield, protein content and protein quality by multi-trait genomic selection in bread wheat. Theor Appl Genet 132:2767–2780
Michel S, Löschenberger F, Ametz C, Pachler B, Sparry E, Bürstmayr H (2019b) Simultaneous selection for grain yield and protein content in genomics-assisted wheat breeding. Theor Appl Genet 132:1745–1760
Michel S, Löschenberger F, Hellinger J, Strasser V, Ametz C, Pachler B, Sparry E, Bürstmayr H (2019c) Improving and maintaining winter hardiness and frost tolerance in bread wheat by genomic selection. Front Plant Sci 10:1195
Murphy KM, Campbell KG, Lyon SR, Jones SS (2007) Evidence of varietal adaptation to organic farming systems. Field Crops Res 102:172–177
Pérez P, De Los CG (2014) Genome-wide regression and prediction with the BGLR statistical package. Genetics 198:483–495
Perez-Lara E, Semagn K, Chen H, Iqbal M, N’Diaye A, Kamran A, Navabi A, Pozniak C, Spaner D (2016) QTLs Associated with agronomic traits in the Cutler × AC Barrie spring wheat mapping population using single nucleotide polymorphic markers. PLoS ONE 11:e0160623
Perez-Lara E, Semagn K, Tran AN, Ciechanowska I, Chen H, Iqbal M, N’Diaye A, Pozniak C, Strelkov SE, Hucl PJ, Graf RJ, Randhawa H, Spaner D (2017) Population structure and genomewide association analysis of resistance to disease and insensitivity to Ptr toxins in Canadian spring wheat using 90K SNP array. Crop Sci 57:1522–1539
Platten JD, Cobb JN, Zantua RE (2019) Criteria for evaluating molecular markers: comprehensive quality metrics to improve marker-assisted selection. PLoS ONE 14:e0210529
Poland J, Endelman J, Dawson J, Rutkoski J, Wu SY, Manes Y, Dreisigacker S, Crossa J, Sanchez-Villeda H, Sorrells M, Jannink JL (2012) Genomic selection in wheat breeding using genotyping-by-sequencing. Plant Genome 5:103–113
Raftery AE, Lewis SM (1992) One long run with diagnostics: Implementation strategies for markov chain monte carlo. Stat Sci 7:493–497
Reid TA, Yang RC, Salmon DF, Navabi A, Spaner D (2011) Realized gains from selection for spring wheat grain yield are different in conventional and organically managed systems. Euphytica 177:253–266
Roorkiwal M, Jarquin D, Singh MK, Gaur PM, Bharadwaj C, Rathore A, Howard R, Srinivasan S, Jain A, Garg V, Kale S, Chitikineni A, Tripathi S, Jones E, Robbins KR, Crossa J, Varshney RK (2018) Genomic-enabled prediction models using multi-environment trials to estimate the effect of genotype × environment interaction on prediction accuracy in chickpea. Sci Rep 8:11701
Rutkoski JE, Heffner EL, Sorrells ME (2011) Genomic selection for durable stem rust resistance in wheat. Euphytica 179:161–173
Rutkoski J, Singh RP, Huerta-Espino J, Bhavani S, Poland J, Jannink JL, Sorrells ME (2015) Efficient use of historical data for genomic selection: a case study of stem rust resistance in wheat. Plant Genome 8:1–10
Schaid DJ, Chen W, Larson NB (2018) From genome-wide associations to candidate causal variants by statistical fine-mapping. Nat Rev Genet 19:491–504
Semagn K, Iqbal M, Chen H, Perez-Lara E, Bemister DH, Xiang R, Zou J, Asif M, Kamran A, N’Diaye A, Randhawa H, Beres BL, Pozniak C, Spaner D (2021a) Physical mapping of QTL associated with agronomic and end-use quality traits in spring wheat under conventional and organic management systems. Theor Appl Genet. https://doi.org/10.1007/s00122-021-03923-x
Semagn K, Iqbal M, Chen H, Perez-Lara E, Bemister DH, Xiang R, Zou J, Asif M, Kamran A, N’Diaye A, Randhawa H, Pozniak C, Spaner D (2021b) Physical mapping of QTL in four spring wheat populations under conventional and organic management systems. I Earl Plants 10:853
Song J, Carver BF, Powers C, Yan L, Klápště J, El-Kassaby YA, Chen C (2017) Practical application of genomic selection in a doubled-haploid winter wheat breeding program. Mol Breed 37:117
Sukumaran S, Crossa J, Jarquin D, Lopes M, Reynolds MP (2017) Genomic prediction with pedigree and genotype × environment interaction in spring wheat grown in South and West Asia, North Africa, and Mexico. Genes Genom Genet 7:481–495
Sukumaran S, Jarquin D, Crossa J, Reynolds M (2018) Genomic-enabled prediction accuracies increased by modeling genotype × environment interaction in Durum wheat. Plant Genome 11:170112
Thavamanikumar S, Dolferus R, Thumma BR (2015) Comparison of genomic selection models to predict flowering time and spike grain number in two hexaploid wheat doubled haploid populations. Genes Genom Genet 5:1991–1998
Thorwarth P, Piepho HP, Zhao Y, Ebmeyer E, Schacht J, Schachschneider R, Kazman E, Reif JC, Würschum T, Longin CFH (2018) Higher grain yield and higher grain protein deviation underline the potential of hybrid wheat for a sustainable agriculture. Plant Breed 137:326–337
VanRaden PM (2008) Efficient methods to compute genomic predictions. J Dairy Sci 91:4414–4423
Veenstra LD, Poland J, Jannink JL, Sorrells ME (2020) Recurrent genomic selection for wheat grain fructans. Crop Sci 60:1499–1512
Vivek BS, Krishna GK, Vengadessan V, Babu R, Zaidi PH, Kha LQ, Mandal SS, Grudloyma P, Takalkar S, Krothapalli K, Singh IS, Ocampo ETM, Xingming F, Burgueño J, Azrai M, Singh RP, Crossa J (2007) Crossa J (2017) Use of genomic estimated breeding values results in rapid genetic gains for drought tolerance in maize. Plant Genome 10(plantgenome2016):0070
Wang S, Wong D, Forrest K, Allen A, Chao S, Huang BE, Maccaferri M, Salvi S, Milner SG, Cattivelli L, Mastrangelo AM, Whan A, Stephen S, Barker G, Wieseke R, Plieske J, Lillemo M, Mather D, Appels R, Dolferus R, Brown-Guedira G, Korol A, Akhunova AR, Feuillet C, Salse J, Morgante M, Pozniak C, Luo MC, Dvorak J, Morell M, Dubcovsky J, Ganal M, Tuberosa R, Lawley C, Mikoulitch I, Cavanagh C, Edwards KJ, Hayden M, Akhunov E (2014) Characterization of polyploid wheat genomic diversity using a high-density 90,000 single nucleotide polymorphism array. Plant Biotechnol J 12:787–796
Wolfe MS, Baresel JP, Desclaux D, Goldringer I, Hoad S, Kovacs G, Löschenberger F, Miedaner T, Østergård H, Lammerts van Bueren ET (2008) Developments in breeding cereals for organic agriculture. Euphytica 163:323
Xiang R, Semagn K, Iqbal M, Chen H, Yang R-C, Spaner D (2021) Phenotypic performance and associated QTL of ‘Peace’ × ‘CDC Stanley’ mapping population under conventional and organic management systems. Crop Sci 61:3469–3483
Xu Y, Wang R, Tong Y, Zhao H, Xie Q, Liu D, Zhang A, Li B, Xu H, An D (2014) Mapping QTLs for yield and nitrogen-related traits in wheat: influence of nitrogen and phosphorus fertilization on QTL expression. Theor Appl Genet 127:59–72
Zhang X, Pérez-Rodríguez P, Burgueño J, Olsen M, Buckler E, Atlin G, Prasanna BM, Vargas M, San Vicente F, Crossa J (2017) Rapid cycling genomic selection in a multiparental tropical maize population. Genes Genom Genet 7:2315–2326
Zhang J, Wang Y, Zhao Y, Zhang Y, Zhang J, Ma H, Han Y (2020) Transcriptome analysis reveals nitrogen deficiency induced alterations in leaf and root of three cultivars of potato (Solanum tuberosum L.). PLoS ONE 15:e0240662
Zhou L, Mrode R, Zhang S, Zhang Q, Li B, Liu J-F (2018) Factors affecting GEBV accuracy with single-step Bayesian models. Heredity 120:100–109
Zou J, Semagn K, Iqbal M, N’Diaye A, Chen H, Asif M, Navabi A, Perez-Lara E, Pozniak C, Yang RC, Randhawa H, Spaner D (2017a) Mapping QTLs controlling agronomic traits in the Attila x CDC Go spring wheat population under organic management using 90K SNP array. Crop Sci 57:365–377
Zou J, Semagn K, Iqbal M, N’Diaye A, Chen H, Asif M, Navabi A, Perez-Lara E, Pozniak C, Yang RC, Randhawa H, Spaner D (2017b) QTLs associated with agronomic traits in the Attila × CDC Go spring wheat population evaluated under conventional management. PLoS ONE 12:e0171528
Acknowledgements
The authors would also like to express appreciation to Klaus Strenzke, Joseph Moss, Izabela Ciechnowska, Fabiana Dias, Katherine Chabot, Tom Keady, and Russel Puk for their technical support with phenotypic data collection in both the conventional and organic managements.
Funding
This study was supported by grants to the University of Alberta wheat breeding program from the Alberta Crop Industry Development Fund (ACIDF), Alberta Wheat Commission (AWC), Saskatchewan Wheat Development Commission (Sask Wheat), Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery and Collaborative Grant, Agriculture and Agri-Food Canada (AAFC), Western Grains Research Foundation Endowment Fund (WGRF), and Core Program Check-off funds to DS.
Author information
Authors and Affiliations
Contributions
KS conceptualized the work, curated and analyzed the data, and wrote the paper. MI supervised and managed the field data collection, contributed to data curation and analyses, and edited the paper. JC, DR, and RH were involved in data analyses and edited the paper. HC, DHB, HR, BLB, AN, and CP were involved in data generation and edited the paper. DS conceptualized the project, acquired funding, supervised the project, and edited the paper.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Availability of data and material
All relevant files are included in this article and its supplementary files.
Code availability
Not applicable.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Communicated by Xianchun Xia.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Semagn, K., Iqbal, M., Crossa, J. et al. Genome-based prediction of agronomic traits in spring wheat under conventional and organic management systems. Theor Appl Genet 135, 537–552 (2022). https://doi.org/10.1007/s00122-021-03982-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-021-03982-0