Abstract
Key message
A key candidate gene for maize kernel length was fine mapped to an interval of 942 kb; the locus significantly increases kernel length (KL) and hundred-kernel weight (HKW).
Abstract
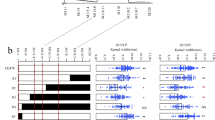
Kernel size is a major determinant of yield in cereals. Kernel length, one of the determining factors of kernel size, is a target trait for both domestication and artificial breeding. However, there are few reports of fine mapping and quantitative trait loci (QTLs)/cloned genes for kernel length in maize. In this project, a novel major QTL, named qKL9, controlling maize kernel length was identified. We verified the authenticity and stability of qKL9 via BC2F2 and BC3F1 populations, respectively, and ultimately mapped qKL9 to an ~ 942-kb genomic interval by testing the progenies of recombination events derived from BC3F2 and BC4F2 populations in multiple environments. Additionally, one new line (McqKL9−A) containing the ~ 942-kb segment was screened from the BC4F2 population. Combining transcriptome analysis between McqKL9−A and Mc at 6, 9 and 14 days after pollination and candidate regional association mapping, Zm00001d046723 was preliminarily identified as the key candidate gene for qKL9. Importantly, the replacement in the Mc line of the Mc’s alleles by the V671’s alleles in the qKL9 region improved the performances of single-cross hybrids obtained with elite lines, illustrating the potential value of this QTL for the genetic improvement in maize kernel-related traits. These findings facilitate molecular breeding for kernel size and cloning of the gene underlying qKL9, shedding light on the genetic basis of kernel size in maize.






Similar content being viewed by others
References
Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11:R106
Becraft PW, Stinard PS, McCarty DR (1996) Crinkyl4: A TNFR-like receptor kinase involved in maize epidermal differentiation. Science 273:1406
Bernardi J, Li Q, Gao Y, Zhao Y, Battaglia R, Marocco A, Chourey PS (2016) The auxin-deficient defective kernel18 (dek18) mutation alters the expression of seed-specific biosynthetic genes in maize. J Plant Growth Regul 35:770–777
Bortiri E, Jackson D, Hake S (2006) Advances in maize genomics: the emergence of positional cloning. Curr Opin Plant Biol 9(2):164–171
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23(19):2633–2635
Buckler ES, Gaut BS, McMullen MD (2006) Molecular and functional diversity of maize. Curr Opin Plant Biol 9(2):172–176
Cai M, Li S, Sun C, Feng Y, Sun Q, Zhao H, Ren X, Zhao Y, Tan B, Qiu F (2017) Emp10 encodes a mitochondrial PPR protein that affects the cis-splicing of nad2 intron 1 and seed development in maize. Plant J 91(1):132–144
Chen J, Zeng B, Zhang M, Xie S, Wang G, Hauck A, Lai J (2014) Dynamic transcriptome landscape of maize embryo and endosperm development. Plant Physiol 166:252–264
Chen L, Li Y, Li C, Wu X, Qin W, Li X, Jiao F, Zhang X, Zhang D, Shi Y, Song Y, Li Y, Wang T (2016) Fine-mapping of qGW4.05, a major QTL for kernel weight and size in maize. BMC Plant Biol 16:81
Chen J, Zhang L, Liu S, Li Z, Huang R, Li Y (2016) The genetic basis of natural variation in kernel size and related traits using a four-way cross population in maize. PLoS ONE 11(4):e0153428. https://doi.org/10.1371/journal.pone.0153428
Chen X, Feng F, Qi W, Xu L, Yao D, Wang Q, Song R (2017) Dek35 encodes a PPR protein that affects cis-splicing of mitochondrial nad4 intron 1 and seed development in maize. Mol Plant 10:427–441
Chen L, Li YX, Li C, Shi Y, Song Y, Zhang D, Wang H, Li Y, Wang T (2020) The retromer protein ZmVPS29 regulates maize kernel morphology likely through an auxin-dependent process(es). Plant Biotechnol J 18(4):1004–1014
Cheng WH, Taliercio WE, Chourey PS (1996) The míniaturel seed locus of maize encodes a cell wali lnvertase required for normal development of endosperm and maternal cells in the pedicel. Plant Cell 8:971–983
Cosgrove DJ (2000) Loosening of plant cell walls by expansins. Nature 407:321–326
Dong X, Ji J, Yang L et al (2019) Fine-mapping and transcriptome analysis of BoGL-3, a wax-less gene in cabbage (Brassica oleracea L. var. capitata). Mol Genet Genomics 294(5):1231–1239. https://doi.org/10.1007/s00438-019-01577-5
Elomaa P, Mehto M, Kotilainen M, Helariutta Y, Nevalainen L, Teeri TH (1998) A bHLH transcription factor mediates organ, region and flower type specific signals on dihydroflavonol-4-reductase (dfr) gene expression in the inflorescence of Gerbera hybrida (Asteraceae). Plant J 16:93–99
Fan C, Xing Y, Mao H, Lu T, Han B, Xu C, Li X, Zhang Q (2006) GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor Appl Genet 112:1164–1171
Feuillet C, Eversole K (2009) Solving the maze. Science 326(5956):1071–1072
Fu S, Meeley R, Scanlon MJ (2002) Empty pericarp2 encodes a negative regulator of the heat shock response and is required for maize embryogenesis. Plant Cell 14:3119–3132
Gutierrez-Marcos JF, Dal Pra M, Giulini A, Costa LM, Gavazzi G, Cordelier S, Sellam O, Tatout C, Paul W, Perez P, Dickinson HG, Consonni G (2007) Empty pericarp4 encodes a mitochondrion-targeted pentatricopeptide repeat protein necessary for seed development and plant growth in maize. Plant Cell 19:196–210
Heang D, Sassa H (2012) An atypical bHLH protein encoded by positive regulator of grain length 2 is involved in controlling grain length and weight of rice through interaction with a typical bHLH protein APG. Breed Sci 62:133–141
Helariutta Y, Elomaa P, Kotilainen M, Seppänen P, Teeri TH (1993) Cloning of cDNA coding for dihydroflavonol-4-reductase (DFR) and characterization of dfr expression in the corollas of Gerbera hybrida var. Regina (Compositae). Plant Mol Biol 22:183–193
Hiebert CW, Moscou MJ, Hewitt T et al (2020) Stem rust resistance in wheat is suppressed by a subunit of the mediator complex. Nature Communication 11(1):1123. https://doi.org/10.1038/s41467-020-14937-2
Hoshino A, Johzuka-Hisatomi Y, Iida S (2001) Gene duplication and mobile genetic elements in the morning glories. Gene 265:1–10
Hu Z, He H, Zhang S, Sun F, Xin X, Wang W, Qian X, Yang J, Luo X (2012) A Kelch motif-containing serine/threonine protein phosphatase determines the large grain QTL trait in rice. J Integr Plant Biol 54:979–990
Huang R, Jiang L, Zheng J, Wang T, Wang H, Huang Y, Hong Z (2013) Genetic bases of rice grain shape: so many genes, so little known. Trends Plant Sci 18:218–226
Inagaki Y, Johzuka-Hisatomi Y, Mori T, Takahashi S, Hayakawa Y, Peyachoknagul S, Ozeki Y, Iida S (1999) Genomic organization of the genes encoding dihydroflavonol 4-reductase for flower pigmentation in the Japanese and common morning glories. Gene 226:181–188
Ishimaru K, Hirotsu N, Madoka Y, Murakami N, Hara N, Onodera H, Kashiwagi T, Ujiie K, Shimizu B, Onishi A, Miyagawa H, Katoh E (2013) Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nat Genet 45:707–711
Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14:R36
Li Q, Li L, Yang X, Warburton ML, Bai G, Dai J, Li J, Yan J (2010) Relationship, evolutionary fate and function of two maize co-orthologs of rice GW2 associated with kernel size and weight. BMC Plant Biol 10:143
Li Q, Yang X, Bai G, Warburton ML, Mahuku G, Gore M, Dai J, Li J, Yan J (2010) Cloning and characterization of a putative GS3 ortholog involved in maize kernel development. Theor Appl Genet 120:753–763
Li Y, Fan C, Xing Y, Jiang Y, Luo L, Sun L, Shao D, Xu C, Li X, Xiao J, He Y, Zhang Q (2011) Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat Genet 43:1266–1269
Li Q, Yang XH, Xu ST, Cai Y, Zhang DL, Han YJ, Li L (2012) Genome-wide association studies identified three independent polymorphisms associated with alpha-tocopherol content in maize kernels. PLoS ONE 7:e36807
Li H, Peng Z, Yang X, Wang W, Fu J, Wang J, Han Y, Chai Y, Guo T, Yang N, Liu J, Warburton ML, Cheng Y, Hao X, Zhang P, Zhao J, Liu Y, Wang G, Li J, Yan J (2013) Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels. Nat Genet 45:43–50
Li C, Li Y, Sun B, Peng B, Liu C, Liu Z, Yang Z, Li Q, Tan W, Zhang Y (2013) Quantitative trait loci mapping for yield components and kernel-related traits in multiple connected RIL populations in maize. Euphytica 193:303–316
Li Y, Lili Tu, Pettolino FA, Ji S, Hao J, Yuan D, Deng F, Tan J, Haiyan Hu, Wang Q, Llewellyn DJ, Zhang X (2016) GbEXPATR, a species-specific expansin, enhances cotton fibre elongation through cell wall restructuring. Plant Biotechnol J 14:951–963
Lid SE, Gruis D, Jung R, Lorentzen JA, Ananiev E, Chamberlin M, Niu X, Meeley R, Nichols S, Olsen OA (2002) The defective kernel 1 (dek1) gene required for aleurone cell development in the endosperm of maize grains encodes a membrane protein of the calpain gene superfamily. Proc Natl Acad Sci USA 99:5460–5465
Liu Y, Wang L, Sun C, Zhang Z, Zheng Y, Qiu F (2014) Genetic analysis and major QTL detection for maize kernel size and weight in multi-environments. Theor Appl Genet 127:1019–1037
Liu J, Deng M, Guo H, Raihan S, Luo J, Xu Y, Dong X, Yan J (2015) Maize orthologs of rice GS5 and their trans-regulator are associated with kernel development. J Integr Plant Biol 57:943–953
Liu D, Zhang J, Liu X et al (2016) Fine mapping and RNA-Seq unravels candidate genes for a major QTL controlling multiple fiber quality traits at the T1 region in upland cotton. BMC Genomics 17:295. https://doi.org/10.1186/s12864-016-2605-6
Liu J, Huang J, Guo H, Lan L, Wang H, Li Q, Yan J et al (2017) The conserved and unique genetic architecture of kernel size and weight in maize and rice. Plant Physiol 175(2):774–785
Liu H, Ren D, Jiang L et al (2020) A natural variation in pleiotropic developmental defects uncovers a crucial role for chloroplast tRNA modification in translation and plant development. Plant Cell. https://doi.org/10.1105/tpc.19.00660
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4325
Panaud O, Chen X, McCouch SR (1996) Development of microsatellite markers and characterization of simple sequence length polymorphism (SSLP) in rice (Oryza sativa L.). Mol Gen Genet 252:597–607
Raihan MS, Liu J, Huang J, Guo H, Pan Q, Yan J (2016) Multi-environment QTL analysis of grain morphology traits and fine mapping of a kernel-width QTL in Zheng58 x SK maize population. Theor Appl Genet 129:1465–1477
Scanlon MJ, Myers AM (1998) Phenotypic analysis and molecular cloning of discolored-1 (dsc1), amaize gene required for early kernel development. Plant Mol Biol 37:483–493
Shen B, Li C, Min Z, Meeley RB, Mc T, Olsen OA (2003) Sal1 determines the number of aleurone cell layers in maize endosperm and encodes a class E vacuolar sorting protein. Proc Natl Acad Sci USA 100:6552–6557
Shirley BW, Hanley S, Goodman HM (1992) Effects of ionizing radiation on a plant genome: analysis of two Arabidopsis transparent testa mutations. Plant Cell 4:333–347
Si L, Chen J, Huang X, Gong H, Luo J, Hou Q, Zhou T, Lu T, Zhu J, Shangguan Y, Chen E, Gong C, Zhao Q, Jing Y, Zhao Y, Li Y, Cui L, Fan D, Lu Y, Weng Q (2016) OsSPL13 controls grain size in cultivated rice. Nat Genet 48:447–456
Song XJ, Huang W, Shi M, Zhu MZ, Lin HX (2007) A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat Genet 39:623–630
Sun L, Li X, Fu Y, Zhu Z, Tan L, Liu F, Sun X, Sun X, Sun C (2013) GS6, a member of the GRAS gene family, negatively regulates grain size in rice. J Integr Plant Biol 55:938–949
Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7:562–578
Wang G, Gao Y, Wang J, Yang L, Song R, Li X, Shi J (2011) Overexpression of two cambium-abundant Chinese fir (Cunninghamia lanceolata) α-expansin genes ClEXPA1 and ClEXPA2 affect growth and development in transgenic tobacco and increase the amount of cellulose in stem cell walls. Plant Biotechnol J 9:486–502
Wang JK, Li HH, Zhang LY, Meng L (2012) QTL IciMapping version 3.2. The Quantitative Genetics Group Institute of Crop Science Chinese Academy of Agricultural Sciences CAAS, Beijing
Wang S, Wu K, Yuan Q, Liu X, Liu Z, Lin X, Zeng R, Zhu H, Dong G, Qian Q, Zhang G, Fu X (2012) Control of grain size, shape and quality by OsSPL16 in rice. Nat Genet 44:950–954
Wang S, Li S, Liu Q, Wu K, Zhang J, Wang S, Wang Y, Chen X, Zhang Y, Gao C, Wang F, Huang H, Fu X (2015) The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat Genet 47:949–954
Wang Y, Xiong G, Hu J, Jiang L, Yu H, Xu J, Fang Y, Zeng L, Xu E, Xu J, Ye W, Meng X, Liu R, Chen H, Jing Y, Wang Y, Zhu X, Li J, Qian Q (2015) Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat Genet 47:944–948
Weng J, Gu S, Wan X, Gao H, Guo T, Su N, Lei C, Zhang X, Cheng Z, Guo X, Wang J, Jiang L, Zhai H, Wan J (2008) Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Res 18:1199–1209
Wen J, Jiang F, Weng Y et al (2019) Identification of heat-tolerance QTLs and high-temperature stress-responsive genes through conventional QTL mapping, QTL-seq and RNA-seq in tomato. BMC Plant Biol 19(1):398. https://doi.org/10.1186/s12870-019-2008-3
Xie D-Y, Jackson LA, Cooper JD, Ferreira D, Paiva NL (2004) Molecular and biochemical analysis of two cDNA clones encoding dihydroflavonol-4-reductase from Medicago truncatula. Plant Physiol 134:979–994
Yang W, Guo Z, Huang C, Duan L, Chen G, Jiang N, Fang W, Feng H, Xie W, Lian X, Wang G (2014) Combining high-throughput phenotyping and genome-wide association studies to reveal natural genetic variation in rice. Nat Commun 5:5087
Yang N, Liu J, Gao Q, Songtao Gui L, Chen LY, Huang J, Deng T, Luo J, He L, Wang Y, Pengwei X, Peng Y, Shi Z, Lan L, Ma Z, Yang X, Zhang Q, Bai M, Li S, Li W, Liu L, Jackson D, Yan J (2019) Genome assembly of a tropical maize inbred line provides insights into structural variation and crop improvement. Nat Genet 51(6):1052–1059
Yi F, Wei Gu, Chen J, Song N, Gao X, Zhang X, Zhou Y, Ma X, Song W, Zhao H, Esteban E, Pasha A, Provart NJ, Lai J (2019) High temporal-resolution transcriptome landscape of early maize seed development. Plant Cell 31(5):974–992
Yu Z, Kang B, He XW, Lv SL, Bai YH, Ding WN, Chen M, Cho H-T, Wu P (2011) Root hair-specific expansins modulate root hair elongation in rice. Plant Biotechnol J 66:725–734
Zhang Z, Liu Z, Hu Y, Li W, Fu Z, Ding D, Li H, Qiao M, Tang J (2014) QTL analysis of kernel-related traits in maize using an immortalized F2 population. PLoS ONE 9:e89645
Zhang J, Sun H, Guo S et al (2020) Decreased protein abundance of lycopene β-Cyclase contributes to red flesh in domesticated watermelon. Plant Physiol. https://doi.org/10.1104/pp.19.01409
Zhang K, Su J, Xu M et al (2020) A common wild rice-derived BOC1 allele reduces callus browning in indica rice transformation. Nat Commun 11(1):443. https://doi.org/10.1038/s41467-019-14265-0
Zhao S, Zhao L, Liu F, Wu Y, Zhu Z, Sun C, Tan L (2016) Narrow and rolled leaf 2 regulates leaf shape, male fertility, and seed size in rice. J Integr Plant Biol 58:983–996
Zhengbin L, Arturo G, Michael DM, Sherry A (2016) Genetic analysis of kernel traits in maize-teosinte introgression populations. G3 6(8):2523–2530
Acknowledgements
This work was supported by the National Key Research and Development Program of China (2016YFD0101803 and 2018YFD0100100) and the National Natural Science Foundation of China (31771796). All authors have read and agreed with the content of the manuscript.
Author information
Authors and Affiliations
Contributions
FQ and DG designed the research; DG and ZT performed the experiments and analyzed the data; HZ analyzed the data; ZP and QS conducted the field trials; DG and FQ wrote the manuscript and all authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Laurence Moreau.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Gong, D., Tan, Z., Zhao, H. et al. Fine mapping of a kernel length-related gene with potential value for maize breeding. Theor Appl Genet 134, 1033–1045 (2021). https://doi.org/10.1007/s00122-020-03749-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-020-03749-z