Abstract
Key message
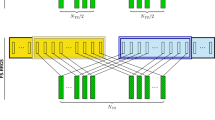
Simulations showed that hybrid performances issued from an incomplete factorial between segregating families of two heterotic groups enable to calibrate genomic predictions of hybrid value more efficiently than tester-based designs.
Abstract
Genomic selection offers new opportunities to revisit hybrid breeding by replacing extensive phenotyping of hybrid combinations by genomic predictions. A key question remains to identify the best design to calibrate genomic prediction models. We proposed to use single-cross hybrids issued from an incomplete factorial design between segregating populations and compared this strategy with a conventional approach based on topcross evaluation. Two multiparental segregating populations of lines, each specific of one heterotic group, were simulated. Hybrids considered as training sets were generated using either (1) a parental line from the opposite group as tester or (2) following an incomplete factorial design. Different specific combining ability (SCA) proportions were simulated by considering different levels of group divergence and dominance effects for the simulated QTL. For the incomplete factorial design, for a same number of hybrids, we considered different numbers of parental lines and different contributions of lines (one to four) to calibration hybrids. We evaluated for different training set sizes prediction accuracies of new hybrids and genetic gains along three generations. At a given training set size, factorial design was as efficient (considering accuracy) as tester design in additive scenarios, but significantly outperformed tester design when SCA was present. The contribution number of each parental line to the incomplete factorial design had a small impact on accuracies. Our simulations confirmed experimental results and showed that calibrating models on hybrids between two multiparental populations is a cost-efficient way to perform genomic predictions in both groups, opening prospects for revisiting reciprocal recurrent selection schemes.






Similar content being viewed by others
References
Akdemir D, Sanchez JI, Jannink J-L (2015) Optimization of genomic selection training populations with a genetic algorithm. Genet Sel Evolut 47:38
Albrecht T, Wimmer V, Auinger H-J, Erbe M, Knaak C et al (2011) Genome-based prediction of testcross values in maize. Theor Appl Genet 123:339–350
Allier A, Teyssèdre S, Lehermeier C, Claustres B, Maltese S et al (2019) Assessment of breeding programs sustainability: application of phenotypic and genomic indicators to a North European grain maize program. Theor Appl Genet 132:1321–1334
Bauer E, Falque M, Walter H, Bauland C, Camisan C et al (2013) Intraspecific variation of recombination rate in maize. Genome Biol 14:R103
Beckett TJ, Morales AJ, Koehler KL, Rocheford TR (2017) Genetic relatedness of previously Plant-Variety-Protected commercial maize inbreds. PLoS ONE 12:e0189277
Bernardo R (1994) Prediction of maize single-cross performance using RFLPs and information from related hybrids. Crop Sci 34:20–25
Bernardo R (1996) Best linear unbiased prediction of the performance of crosses between untested maize inbreds. Crop Sci 36:872–876
Beyene Y, Semagn K, Mugo S, Tarekegne A, Babu R et al (2015) Genetic gains in grain yield through genomic selection in eight bi-parental maize populations under drought stress. Crop Sci 55:154–163
Blanc G, Charcosset A, Veyrieras J-B, Gallais A, Moreau L (2008) Marker-assisted selection efficiency in multiple connected populations: a simulation study based on the results of a QTL detection experiment in maize. Euphytica 161:71–84
Boeven PHG, Würschum T, Weissmann S, Miedaner T, Maurer HP (2016) Prediction of hybrid performance for fusarium head blight resistance in triticale (× triticosecale wittmack). Euphytica 207:475–490
Browning SR, Browning BL (2007) Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet 81:1084–1097
Coor JG (1997) Selection methodologies and heterosis. CIMMYT. Book of Abstracts p170. The genetics and exploitation of heterosis in crops; an international symposium, Mexico, D.F., Mexico
Crossa J, Pérez-Rodríguez P, Cuevas J, Montesinos-López O, Jarquín D et al (2017) Genomic selection in plant breeding: methods, models, and perspectives. Trends Plant Sci 22:961–975
Fritsche-Neto R, Akdemir D, Jannink J-L (2018) Correction to: accuracy of genomic selection to predict maize single-crosses obtained through different mating designs. Theor Appl Genet 131:1603–1603
Ganal MW, Durstewitz G, Polley A, Bérard A, Buckler ES et al (2011) A large maize (Zea mays L.) SNP genotyping array: development and germplasm genotyping, and genetic mapping to compare with the B73 reference genome. PLoS ONE. https://doi.org/10.1371/journal.pone.0028334
Giraud H, Lehermeier C, Bauer E, Falque M, Segura V, Bauland C et al (2014) Linkage disequilibrium with linkage analysis of multiline crosses reveals different multiallelic QTL for hybrid performance in the flint and dent heterotic groups of maize. Genetics 198:1717–1734
Giraud H, Bauland C, Falque M, Madur D, Combes V et al (2017) Reciprocal genetics: identifying QTL for general and specific combining abilities in hybrids between multiparental populations from two maize (Zea mays L.) heterotic groups. Genetics 207:1167–1180
Giraud H (2016) Genetic analysis of hybrid value for silage maize in multiparental designs: QTL detection and genomic selection. PhD thesis. Université Paris-Saclay. https://tel.archives-ouvertes.fr/tel-01443275
Gore MA, Chia J-M, Elshire RJ, Sun Q, Ersoz ES et al (2009) A first-generation haplotype map of maize. Science 326:1115–1117
Gowda M, Zhao Y, Maurer HP, Weissmann EA, Würschum T et al (2013) Best linear unbiased prediction of triticale hybrid performance. Euphytica 191:223–230
Hallauer AR, Eberhart SA (1970) Reciprocal full-sib selection. Crop Sci 10(3):315–316
Hallauer AR, Filho M, Carena MJ (2010) Quantitative genetics in maize breeding. In: Rajcan I, Vollmann J (eds) Handbook of plant breeding. Springer, New York
Henderson CR (1973) Sire evaluation and genetic trends. J Anim Sci 1973:10–41
Hofheinz N, Borchardt D, Weissleder K, Frisch M (2012) Genome-based prediction of test cross performance in two subsequent breeding cycles. Theor Appl Genet 125:1639–1645
Jacobson A, Lian L, Zhong SQ, Bernardo R (2014) General Combining Ability model for genomewide selection in a biparental cross. Crop Sci 54(3):895–905
Kadam DC, Lorenz AJ (2018) Toward redesigning hybrid maize breeding through genomics-assisted breeding in the maize genome. In: Bennetzen J, Flint-Garcia S, Hirsch C, Tuberosa R (eds) Compendium of plant genomes. Springer, Cham, pp 367–88
Kadam DC, Lorenz AJ (2019) Evaluation of nonparametric models for genomic prediction of early-stage single crosses in maize. Crop Sci 59(4):1411–1423
Kadam DC, Potts SM, Bohn MO, Lipka AE, Lorenz AJ (2016) Genomic prediction of single crosses in the early stages of a maize hybrid breeding pipeline. G3 6:3443–3453
Laporte F, Mary-Huard T (2018) MM4LMM: inference of linear mixed models through MM algorithm (version 1.0.5). https://CRAN.R-project.org/package=MM4LMM. Accessed 31 May 2018
Longin CFH, Mi X, Würschum T (2015) Genomic selection in wheat: optimum allocation of test resources and comparison of breeding strategies for line and hybrid breeding. Theor Appl Genet 128:1297–1306
Maenhout S, De Baets B, Haesaert G, Van Bockstaele E (2007) Support vector machine regression for the prediction of maize hybrid performance. Theor Appl Genet 115:1003–1013
Maenhout S, De Baets B, Haesaert G (2010) Prediction of maize single-cross hybrid performance: support vector machine regression versus best linear prediction. Theor Appl Genet 120:415–427
Mangin B, Bonnafous F, Blanchet N et al (2017) Genomic prediction of sunflower hybrids oil content. Front Plant Sci 8:1633. https://doi.org/10.3389/fpls.2017.01633
Marulanda JJ, Mi X, Melchinger AE, Xu J-L, Würschum T, Longin CFH (2016) Optimum breeding strategies using genomic selection for hybrid breeding in wheat, maize, rye, barley, rice and triticale. Theor Appl Genet 129:1901–1913
Massman JM, Gordillo A, Lorenzana RE, Bernardo R (2013a) Genomewide predictions from maize single-cross data. Theor Appl Genet 126:13–22
Massman JM, Jung H-JG, Bernardo R (2013b) Genomewide selection versus marker-assisted recurrent selection to improve grain yield and stover-quality traits for cellulosic ethanol in maize. Crop Sci 53:58–66
Melchinger AE, Geiger HH, Seitz G, Scmidt GA (1987) Optimum prediction of three-way crosses from single crosses in forage maize (Zea mays L.). Theor Appl Genet 74(3):339–345
Meuwissen TE, Hayes B, Goddard M (2001) Prediction of total genetic value using genome-wide dense marker maps. Genetics 157:1819–1829
Meuwissen TE, Hayes B, Goddard M (2016) Genomic selection: a paradigm shift in animal breeding. Anim Front 6:6–14
Miedaner T, Zhao Y, Gowda M, Longin CFH, Korzun V, Ebmeyer E, Kazman E, Reif JC (2013) Genetic architecture of resistance to Septoria tritici blotch in European wheat. BMC Genom 14:858
Philipp N, Liu G, Zhao Y, He S, Spiller M et al (2016) Genomic prediction of barley hybrid performance. Plant Genome. https://doi.org/10.3835/plantgenome2016.02.0016
R Core Team (2013) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Reif JC, Gumpert F-M, Fischer S, Melchinger AE (2007) Impact of interpopulation divergence on additive and dominance variance in hybrid populations. Genetics 176:1931–1934
Reif JC, Zhao Y, Würschum T, Gowda M, Hahn V (2013) Genomic prediction of sunflower hybrid performance. Plant Breed 132:107–114
Riedelsheimer C, Endelman JB, Stange M, Sorrells ME, Jannink J-L, Melchinger AE (2013) Genomic predictability of interconnected biparental maize populations. Genetics 194:493–503
Rincent R, Laloë D, Nicolas S, Altmann T, Brunel D, Revilla P, Rodríguez VM et al (2012) Maximizing the reliability of genomic selection by optimizing the calibration set of reference individuals: comparison of methods in two diverse groups of maize inbreds (Zea mays L.). Genetics 192:715–728
Schrag TA, Melchinger AE, Sørensen AP, Frisch M (2006) Prediction of single-cross hybrid performance for grain yield and grain dry matter content in maize using AFLP markers associated with QTL. Theor Appl Genet 113:1037–1047
Schrag TA, Frisch M, Dhillon BS, Melchinger AE (2009) Marker-based prediction of hybrid performance in maize single-crosses involving doubled haploids. Maydica 54:353–362
Schrag TA, Westhues M, Schipprack W, Seifert F, Thiemann A, Scholten S, Melchinger AE (2018) Beyond genomic prediction: combining different types of omics data can improve prediction of hybrid performance in maize. Genetics 208:1373–1385
Seye AI, Bauland C, Giraud H, Mechin V, Reymond M, Charcosset A, Moreau L (2019) Quantitative trait loci mapping in hybrids between dent and flint maize multiparental populations reveals group-specific QTL for silage quality traits with variable pleiotropic effects on yield. Theor Appl Genet 132:1523–1542
Sprague GF, Tatum LA (1942) General vs. specific combining ability in single crosses of corn. Agron J 34:923–932
Technow F, Riedelsheimer C, Schrag TA, Melchinger AE (2012) Genomic prediction of hybrid performance in maize with models incorporating dominance and population specific marker effects. Theor Appl Genet 125:1181–1194
Technow F, Schrag TA, Schipprack W, Bauer E, Simianer H, Melchinger AE (2014) Genome properties and prospects of genomic prediction of hybrid performance in a breeding program of maize. Genetics 197(4):1343–1355
VanRaden PM (2008) Efficient methods to compute genomic predictions. J Dairy Sci 91:4414–4423
Vitezica ZG, Varona L, Elsen J-M, Misztal I, Herring W et al (2016) Genomic BLUP including additive and dominant variation in purebreds and F1 crossbreds, with an application in pigs. Genet Sel Evol 48:6
Wang Y, Mette MF, Miedaner T, Gottwald M, Wilde P et al (2014) The accuracy of prediction of genomic selection in elite hybrid rye populations surpasses the accuracy of marker-assisted selection and is equally augmented by multiple field evaluation locations and test years. BMC Genom 15:556
Werner CR, Qian L, Voss-Fels KP, Abbadi A, Leckband G et al (2018) Genome-wide regression models considering general and specific combining ability predict hybrid performance in oilseed rape with similar accuracy regardless of trait architecture. Theor Appl Genet 131:299–317
Westhues M, Schrag TA, Heuer C, Thaller G, Utz HF et al (2017) Omics-based hybrid prediction in maize. Theor Appl Genet 130:1927–1939
Windhausen VS, Gary NA, Hickey JM, Crossa J, Jannink J-L, Sorrells ME, Raman B et al (2012) Effectiveness of genomic prediction of maize hybrid performance in different breeding populations and environments. G3 2(11):1427–1436
Würschum T, Reif JC, Kraft T, Janssen G, Zhao Y (2013) Genomic selection in sugar beet breeding populations. BMC Genet 14:85
Xu W, Virmani SS, Hernandez JE, Redoña ED, Sebastian LS (2000) Prediction of hybrid performance in rice: comparisons among best linear unbiased prediction (BLUP) procedure, midparent value, and molecular marker distance. Int Rice Res Notes 25:12–13
Xu Y, Wang X, Ding X, Zheng X, Yang Z et al (2018) Genomic selection of agronomic traits in hybrid rice using an NCII population. Rice 11:32
Zhao Y, Zeng J, Fernando R, Reif JC (2013) Genomic prediction of hybrid wheat performance. Crop Sci 53:802–810
Zhao Y, Mette MF, Gowda M, Longin CFH, Reif JC (2014) Bridging the gap between marker-assisted and genomic selection of heading time and plant height in hybrid wheat. Heredity 112:638–645
Zhao Y, Mette MF, Reif JC (2015) Genomic selection in hybrid breeding. Plant Breed 134:1–10
Acknowledgements
A. I. Seye’s PhD was funded by the Senegalese Institute of Agricultural Research (ISRA) through a Scholarship from the West Africa Agricultural Productivity Program (WAAPP) given by the National Institute of Higher Education in Agricultural Sciences—Montpellier SupAgro and the “Amaizing” project (ANR-10-BTBR-0001). We are grateful to Caussade Semences, Corteva Agriscience, Euralis Semences, KWS, Limagrain Europe, Mas Seeds, R2n and Syngenta Seeds grouped in the frame of the ProMais “SAM-MCR” project for the funding. We also thank scientists from these companies for helpful discussions on the results. We thank D. Madur for sharing the CornFed genotyping data.
Author information
Authors and Affiliations
Contributions
CB, AC and LM initiated this project. LM and CB coordinated it. AC and LM supervised this work. AIS wrote the simulation programs, analysed the results and prepared the manuscript. All authors discussed the results and contributed to the final manuscript. All authors revised and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standards
All usual standards have been respected.
Additional information
Communicated by Albrecht E. Melchinger.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Seye, A.I., Bauland, C., Charcosset, A. et al. Revisiting hybrid breeding designs using genomic predictions: simulations highlight the superiority of incomplete factorials between segregating families over topcross designs. Theor Appl Genet 133, 1995–2010 (2020). https://doi.org/10.1007/s00122-020-03573-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-020-03573-5