Abstract
Key message
Next-generation sequencing enabled a fast discovery of QTLs controlling CMV resistant in pepper. The gene CA02g19570 as a possible candidate gene of qCmr2.1 was identified for resistance to CMV in pepper.
Abstract
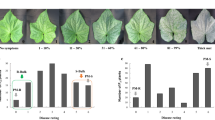
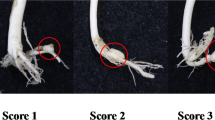
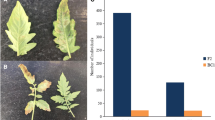
Cucumber mosaic virus (CMV) is one of the most important viruses infecting pepper, but the genetic basis of CMV resistance in pepper is elusive. In this study, we identified a candidate gene for CMV resistance QTL, qCmr2.1 through SLAF-seq. Segregation analysis in F2, BC1 and F2:3 populations derived from a cross between two inbred lines ‘PBC688’ (CMV-resistant) and ‘G29’ (CMV-susceptible) suggested quantitative inheritance of resistance to CMV in pepper. Genome-wide comparison of SNP profiles between the CMV-resistant and CMV-susceptible bulks constructed from an F2 population identified two QTLs, designated as qCmr2.1 on chromosome 2 and qCmr11.1 on chromosome 11 for resistance to CMV in PBC688, which were confirmed by InDel marker-based classical QTL mapping in the F2 population. As a major QTL, joint SLAF-seq and traditional QTL analysis delimited qCmr2.1 to a 330 kb genomic region. Two pepper genes, CA02g19570 and CA02g19600, were identified in this region, which are homologous with the genes LOC104113703, LOC104248995, LOC102603934 and LOC101248357, which were predicted to encode N-like protein associated with TMV-resistant in Solanum crops. Quantitative RT-PCR revealed higher expression levels of CA02g19570 in CMV resistance genotypes. The CA02g19600 did not exhibit obvious regularity in expression patterns. Higher relative expression levels of CA02g19570 in PBC688 and F1 were compared with those in G29 during days after inoculation. These results provide support for CA02g19570 as a possible candidate gene of qCmr2.1 for resistance to CMV in pepper.







Similar content being viewed by others
References
Abe A, Kosugi S, Yoshida K, Natsume S, Takagi H, Kanzaki H, Matsumura H, Yoshida K, Mitsuoka C, Tamiru M (2012) Genome sequencing reveals agronomically important loci in rice using MutMap. Nat Biotechnol 30:174–178
Austin RS, Vidaurre D, Stamatiou G, Breit R, Provart NJ, Bonetta D, Zhang J, Fung P, Gong Y, Wang PW (2011) Next-generation mapping of Arabidopsis genes. Plant J 67:715–725
Caranta C, Palloix A, Lefebvre V, Daubeze A (1997) QTLs for a component of partial resistance to Cucumber mosaic virus in pepper: restriction of virus installation in host-cells. Theor Appl Genet 94:431–438
Caranta C, Pflieger S, Lefebvre V, Daubeze A, Thabuis A, Palloix A (2002) QTLs involved in the restriction of Cucumber mosaic virus (CMV) long-distance movement in pepper. Theor Appl Genet 104:586–591
Chaim AB, Grube R, Lapidot M, Jahn M, Paran I (2001) Identification of quantitative trait loci associated with resistance to Cucumber mosaic virus in Capsicum annuum. Theor Appl Genet 102:1213–1220
Chen S, Huang Z, Dai Y, Qin S, Gao Y, Zhang L, Gao Y, Chen J (2013) The development of 7E chromosome-specific molecular markers for Thinopyrum elongatum based on SLAF-seq technology. PLoS One. doi:10.1371/journal.pone.0065122
Ehrenreich IM, Torabi N, Jia Y, Kent J, Martis S, Shapiro JA, Gresham D, Caudy AA, Kruglyak L (2010) Dissection of genetically complex traits with extremely large pools of yeast segregants. Nature 464:1039–1042
Green SK, Kim JS (1994) Sources of resistance to viruses of pepper (Capsicum spp.): a catalog. Tech Bull 20:10–19
Grube RC, Radwanski ER, Jahn M (2000a) Comparative genetics of disease resistance within the Solanaceae. Genetics 155:873–887
Grube RC, Zhang Y, Murphy JF, Loaiza-Figueroa F, Lackney VK, Provvidenti R, Jahn MK (2000b) New source of resistance to Cucumber mosaic virus in Capsicum frutescens. Plant Dis 84:885–891
Han Y, Lv P, Hou S, Li S, Ji G, Ma X, Du R, Liu G (2015) Combining next generation sequencing with bulked segregant analysis to fine map a stem moisture locus in Sorghum (Sorghum bicolor L. PloS One, Moench). doi:10.1371/journal.pone.0127065
Hyndman RJ, Fan Y (1996) Sample quantiles in statistical packages. Am Sta 50:361–365
Jacquemond M (2012) Cucumber mosaic virus. Adv Virus Res 84:439–504
Kang W-H, Hoang NH, Yang H-B, Kwon J-K, Jo S-H, Seo J-K, Kim K-H, Choi D, Kang B-C (2010) Molecular mapping and characterization of a single dominant gene controlling CMV resistance in peppers (Capsicum annuum L.). Theor Appl Genet 120:1587–1596
Korol A, Frenkel Z, Cohen L, Lipkin E, Soller M (2007) Fractioned DNA pooling: a new cost-effective strategy for fine mapping of quantitative trait loci. Genetics 176:2611–2623
Lapidot M, Paran I, Ben-Joseph R, Ben-Harush S, Pilowsky M, Cohen S, Shifriss C (1997) Tolerance to Cucumber mosaic virus in pepper: development of advanced breeding lines and evaluation of virus level. Plant Dis 81:185–188
Lefebvre V, Palloix A, Caranta C, Pochard E (1995) Construction of an intraspecific integrated linkage map of pepper using molecular markers and doubled-haploid progenies. Genome 38:112–121
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25:1754–1760
Li R, Li Y, Kristiansen K, Wang J (2008) SOAP: short oligonucleotide alignment program. Bioinformatics 24:713–714
Livaja M, Wang Y, Wieckhorst S, Haseneyer G, Seidel M, Hahn V, Knapp SJ, Taudien S, Schön C-C, Bauer E (2013) BSTA: a targeted approach combines bulked segregant analysis with next-generation sequencing and de novo transcriptome assembly for SNP discovery in sunflower. BMC Genom 14:628–638
Lu H, Lin T, Klein J, Wang S, Qi J, Zhou Q, Sun J, Zhang Z, Weng Y, Huang S (2014) QTL-seq identifies an early flowering QTL located near flowering locus T in cucumber. Theor Appl Genet 127:1491–1499
Michelmore RW, Paran I, Kesseli RV (1991) Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci USA 88:9828–9832
Min W-K, Ryu J-H, Ahn S-H (2014) Developmental changes of recessive genes-mediated Cucumber mosaic virus (CMV) resistance in peppers (Capsicum annuum L.). Kor J Hort Sci Technol 32:235–240
Monma S, Sakata Y (1997) Screening of capsicum accessions for resistance to cucumber mosaic virus. J Jpn Soc Hortic Sci 65:769–776
Murray M, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4326
Pochard E, Dumas de Valuix R, Florent A (1983) Linkage between partial resistance to CMV and susceptibility to TMV in the line Perennial: analysis on androgenetic homozygous lines. Capsicum Eggplant News 2:34–35
Salvi S, Tuberosa R (2005) To clone or not to clone plant QTLs: present and future challenges. Trends Plant Sci 10:297–304
Schneeberger K, Ossowski S, Lanz C, Juul T, Petersen AH, Nielsen KL, Jørgensen J-E, Weigel D, Andersen SU (2009) SHOREmap: simultaneous mapping and mutation identification by deep sequencing. Nat Methods 6:550–551
Stamova BS, Chetelat RT (2000) Inheritance and genetic mapping of Cucumber mosaic virus resistance introgressed from Lycopersicon chilense into tomato. Theor Appl Genet 101:527–537
Sun X, Liu D, Zhang X, Li W, Liu H, Hong W, Jiang C, Guan N, Ma C, Zeng H (2013) SLAF-seq: an efficient method of large-scale de novo SNP discovery and genotyping using high-throughput sequencing. PLoS One. doi:10.1371/journal.pone.0058700
Suzuki K, Kuroda T, Miura Y, Murai J (2003) Screening and field trials of virus resistant sources in Capsicum spp. Plant Dis 87:779–783
Swinnen S, Schaerlaekens K, Pais T, Claesen J, Hubmann G, Yang Y, Demeke M, Foulquié-Moreno MR, Goovaerts A, Souvereyns K (2012) Identification of novel causative genes determining the complex trait of high ethanol tolerance in yeast using pooled-segregant whole-genome sequence analysis. Genome Res 22:975–984
Takagi H, Abe A, Yoshida K, Kosugi S, Natsume S, Mitsuoka C, Uemura A, Utsushi H, Tamiru M, Takuno S (2013) QTL-seq: rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J74:174–183
Takahashi H, Suzuki M, Natsuaki K, Shigyo T, Hino K, Teraoka T, Hosokawa D, Ehara Y (2001) Mapping the virus and host genes involved in the resistance response in cucumber mosaic virus-infected Arabidopsis thaliana. Plant Cell Physiol 42:340–347
Takahashi H, Miller J, Nozaki Y, Takeda M, Shah J, Hase S, Ikegami M, Ehara Y, Dinesh-Kumar S (2002) RCY1, an Arabidopsis thaliana RPP8/HRT family resistance gene, conferring resistance to Cucumber mosaic virus requires salicylic acid, ethylene and a novel signal transduction mechanism. Plant J 32:655–667
Van Ooien J, Boer M, Jansen R, Maliepaard C (2002) MapQTL 4.0. Software for the calculation of QTL positions on genetic maps. Plant Research International, Wageningen
Van Ooijen J (2011) Multipoint maximum likelihood mapping in a full-sib family of an outbreeding species. Genet Res 93:343–349
Wan H, Yuan W, Ruan M, Ye Q, Wang R, Li Z, Zhou G, Yao Z, Zhao J, Liu S (2011) Identification of reference genes for reverse transcription quantitative real-time PCR normalization in pepper (Capsicum annuum L.). Biochem Bioph Res Co 416:24–30
Xu F, Sun X, Chen Y, Huang Y, Tong C, Bao J (2015a) Rapid identification of major QTLs associated with rice grain weight and their utilization. PLoS One. doi:10.1371/journal.pone.0122206
Xu X, Lu L, Zhu B, Xu Q, Qi X, Chen X (2015b) QTL mapping of cucumber fruit flesh thickness by SLAF-seq. Sci Rep UK 5:15829–15838
Yang Z, Huang D, Tang W, Zheng Y, Liang K, Cutler AJ, Wu W (2013) Mapping of quantitative trait loci underlying cold tolerance in rice seedlings via high-throughput sequencing of pooled extremes. PLoS One. doi:10.1371/journal.pone.0068433
Yao M, Li N, Wang F, Ye Z (2013) Genetic analysis and identification of QTLs for resistance to Cucumber mosaic virus in chili pepper (Capsicum annuum L.). Euphytica 193:135–145
Yoshii M, Nishikiori M, Tomita K, Yoshioka N, Kozuka R, Naito S, Ishikawa M (2004) The Arabidopsis cucumovirus multiplication 1 and 2 loci encode translation initiation factors 4E and 4G. J Virol 78:6102–6111
Acknowledgments
This work was supported by the National Natural Science Foundation of China (NSFC 31401866), Natural Science Foundation of Jiangsu Province (CN) (4111415), Jiangsu Provincial Fund for Independent Innovation of Agricultural Sciences [CX (14)5013], and Jiangsu Agriculture Science and Technology Innovation Fund [CX (12)1004].
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have declared that no competing interests exist.
Ethical standard
The experiments in this study comply with the current laws of China.
Additional information
Communicated by M. J. Havey.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Guo, G., Wang, S., Liu, J. et al. Rapid identification of QTLs underlying resistance to Cucumber mosaic virus in pepper (Capsicum frutescens). Theor Appl Genet 130, 41–52 (2017). https://doi.org/10.1007/s00122-016-2790-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-016-2790-3