Abstract
Key message
The APETALA2 transcription factor homolog CaAP2 is a candidate gene for a flowering repressor in pepper, as revealed by induced-mutation phenotype, and a candidate underlying a major QTL controlling natural variation in flowering time.
Abstract
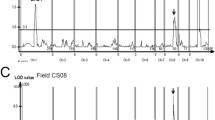
To decipher the genetic control of transition to flowering in pepper (Capsicum spp.) and determine the extent of gene function conservation compared to model species, we isolated and characterized several ethyl methanesulfonate (EMS)-induced mutants that vary in their flowering time compared to the wild type. In the present study, we report on the isolation of an early-flowering mutant that flowers after four leaves on the primary stem compared to nine leaves in the wild-type ‘Maor’. By genetic mapping and sequencing of putative candidate genes linked to the mutant phenotype, we identified a member of the APETALA2 (AP2) transcription factor family, CaAP2, which was disrupted in the early-flowering mutant. CaAP2 is a likely ortholog of AP2 that functions as a repressor of flowering in Arabidopsis. To test whether CaAP2 has an effect on controlling natural variation in the transition to flowering in pepper, we performed QTL mapping for flowering time in a cross between early and late-flowering C. annuum accessions. We identified a major QTL in a region of chromosome 2 in which CaAP2 was the most significant marker, explaining 52 % of the phenotypic variation of the trait. Sequence comparison of the CaAP2 open reading frames in the two parents used for QTL mapping did not reveal significant variation. In contrast, significant differences in expression level of CaAP2 were detected between near-isogenic lines that differ for the flowering time QTL, supporting the putative function of CaAP2 as a major repressor of flowering in pepper.






Similar content being viewed by others
References
Alvarez J, Guli CL, Yu XH, Smyth DR (1992) TERMINAL FLOWER: a gene affecting inflorescence development in Arabidopsis thaliana. Plant J 2:103–116
Ashrafi H, Hill T, Stoffel K, Kozik A, Yao JQ, Chin-Wo SR, Van Deynze A (2012) De novo assembly of the pepper transcriptome (Capsicum annuum): a benchmark for in silico discovery of SNPs, SSRs and candidate genes. BMC Genomics 13:571
Aukerman MJ, Sakai S (2003) Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15:2730–2741
Bowman JL, Smyth DR, Meyerowitz EM (1989) Genes directing flower development in Arabidopsis. Plant Cell 1:37–52
Chung MY, Vrebalov J, Alba R, Lee J, McQuinn R, Chung JD, Klein P, Giovannoni J (2010) A tomato (Solanum lycopersicum) APETALA2/ERF gene, SlAP2a, is a negative regulator of fruit ripening. Plant J 64:936–947
Cohen O, Borovsky Y, David-Schwartz R, Paran I (2012) CaJOINTLESS is a MADS-box gene involved in suppression of vegetative growth in all shoot meristems in pepper. J Exp Bot 63:4947–4957
Cohen O, Borovsky Y, David-Schwartz R, Paran I (2014) Capsicum annuum S (CaS) promotes reproductive transition and is required for flower formation in pepper (Capsicum annuum). New Phytol 202:1014–1023
Corbesier L, Vincent C, Jang S et al (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316:1030–1033
Deng W, Ying H, Helliwell CA, Taylor JM, Peacock WJ, Dennis ES (2011) FLOWERING LOCUS C (FLC) regulates development pathways throughout the life cycle of Arabidopsis. Proc Natl Acad Sci USA 108:6680–6685
Elitzur T, Nahum H, Borovsky Y, Pekker I, Eshed Y, Paran I (2009) Co-ordinated regulation of flowering time, plant architecture and growth by FASCICULATE: the pepper orthologue of SELF PRUNING. J Exp Bot 60:869–880
Golovnina KA, Kondratenko EY, Blinov AG, Goncharov NP (2010) Molecular characterization of vernalization loci VRN1 in wild and cultivated wheats. BMC Plant Biol 10:168
Gul Khan MR, Ai XY, Zhang JZ (2014) Genetic regulation of flowering time in annual and perennial plants. Wiley Interdiscip Rev RNA 5:347–359
Hung H-Y, Shannon LM, Tian F et al (2012) ZmCCT and the genetic basis of day-length adaptation underlying the postdomestication spread of maize. Proc Natl Acad Sci USA 109:E1913–E1921
Jeifetz D, David-Schwartz R, Borovsky Y, Paran I (2011) CaBLIND regulates axillary meristem initiation and transition to flowering in pepper. Planta 234:1227–1236
Jung JH, Lee S, Yun J, Lee M, Park CM (2014) The miR172 target TOE3 represses AGAMOUS expression during Arabidopsis floral patterning. Plant Sci 215–216:29–38
Karlova R, Rosin FM, Busscher-Lange J, Parapunova V, Do PT, Fernie AR, Fraser PD, Baxter C, Angenent GC, deMaagd RA (2011) Transcriptome and metabolite profiling show that APETALA2a is a major regulator of tomato fruit ripening. Plant Cell 23:923–941
Kim S, Park M, Yeom S-I et al (2014) Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat Genet 46:270–278
Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newberg LA (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
Li D, Liu C, Shen L et al (2008) A repressor complex governs the integration of flowering signals in Arabidopsis. Dev Cell 15:110–120
Lu L, Yan W, Xue W, Shao D, Xing Y (2012) Evolution and association analysis of Ghd7 in rice. PLoS One 7:e34021
MacAlister CA, Park SJ, Jiang K, Marcel F, Bendahmane A, Izkovich Y, Eshed Y, Lippman ZB (2012) Synchronization of the flowering transition by the tomato TERMINATING FLOWER gene. Nat Genet 44:1393–1398
Maes T, Van de Steene N, Zethof J, Karimi M, D’Hauw M, Mares G, Van Montagu M, Gerats T (2001) Petunia Ap2-like genes and their role in flower and seed development. Plant Cell 13:229–244
Martin A, Adam H, Diaz-Mendoza M, Zurczak M, Gonzalez-Schain ND, Suarez-Lopez P (2009) Graft-transmissible induction of potato tuberization by the microRNA miR172. Development 136:2873–2881
Mathieu J, Yant LJ, Murdter F, Kuttner F, Schmid M (2009) Repression of flowering by the miR172 target SMZ. PLoS Biol 7:e1000148. doi:10.1371/journal.pbio.1000148
Mendez-Vigo B, Martınez-Zapater JM, Alonso-Blanco C (2013) The flowering repressor SVP underlies a novel Arabidopsis thaliana QTL interacting with the genetic background. PLoS Genet 9:e1003289. doi:10.1371/journal.pgen.1003289
Michaels SD, He Y, Scortecci KC, Amasino RM (2003) Attenuation of FLOWERING LOCUS C activity as a mechanism for the evolution of summer-annual flowering behavior in Arabidopsis. Proc Natl Acad Sci USA 100:10102–10107
Michelmore RW, Paran I, Kesseli RV (1991) Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci USA 88:9828–9832
Nelson CJ (1997) QGENE: software for marker-based genomic analysis and breeding. Mol Breed 3:229–235
Ohto MA, Fischer RL, Goldberg RB, Nakamura K, Harada JJ (2005) Control of seed mass by APETALA2. Proc Natl Acad Sci USA 102:3123–3128
Olsen KM, Wendel JF (2013) A bountiful harvest: genomic insights into crop domestication phenotypes. Ann Rev Plant Biol 64:47–70
Paran I, Borovsky Y, Nahon S, Cohen O (2007) The use of induced mutations to study shoot architecture in Capsicum. Isr J Plant Sci 55:125–131
Park SJ, Jiang K, Schatz MC, Lippman ZB (2012) Rate of meristem maturation determines inflorescence architecture in tomato. Proc Natl Acad Sci USA 109:639–644
Ripoll JJ, Roeder AHK, Ditta GS, Yanofsky MF (2011) A novel role for the floral homeotic gene APETALA2 during Arabidopsis fruit development. Development 138:5167–5176
Rosas U, Mei Y, Xie Q et al (2014) Variation in Arabidopsis flowering time associated with cis-regulatory variation in CONSTANS. Nat Commun. doi:10.1038/ncomms4651
Salomé PA, Bomblies K, Laitinen RAE, Yant L, Mott R, Weigel D (2011) Genetic architecture of flowering-time variation in Arabidopsis thaliana. Genetics 188:421–433
Salvi S, Sponza G, Morgante M et al (2007) Conserved noncoding genomic sequences associated with a flowering-time quantitative trait locus in maize. Proc Natl Acad Sci USA 104:11376–11381
Samach A, Lotan H (2007) The transition to flowering in tomato. Plant Biotechnol 24:71–82
Shrestha R, Ariza JG, Brambilla V, Fornara F (2014) Molecular control of seasonal flowering in rice, arabidopsis and temperate cereals. Ann Bot. doi:10.1093/aob/mcu032
Srikanth A, Schmid M (2011) Regulation of flowering time: all roads lead to Rome. Cell Mol Life Sci 68:2013–2037
Wu F, Eannetta NT, Xu Y, Durrett R, Mazourek M, Jahn MM, Tanksley SD (2009) A COSII genetic map of the pepper genome provides a detailed picture of synteny with tomato and new insights into recent chromosome evolution in the genus Capsicum. Theor Appl Genet 118:1279–1293
Yant L, Mathieu J, Schmid M (2009) Just say no: floral repressors help Arabidopsis bide the time. Curr Opin Plant Biol 12:580–586
Yant L, Mathieu J, Dinh TT, Ott F, Lanz C, Wollmann H, Chen X, Schmid M (2010) Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. Plant Cell 22:2156–2170
Zuellig MP, Kenney AM, Sweigart AL (2014) Evolutionary genetics of plant adaptation: insights from new model systems. Curr Opinion Plant Biol 18:44–50
Acknowledgments
We thank Aharon Bellalou for technical support, Arnon Brand for graphic design and Hanita Zemach for assistance with microscopic analyses. We thank Dr. Zach Lippman (Cold Spring Harbor Laboratory) for contributing the digital expression data of CaAP2 in ‘Maor’. This research was supported by The Israel Science Foundation (Grant No. 1349/10).
Conflict of interest
The authors declare no conflicts of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by J. Dubcovsky.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Borovsky, Y., Sharma, V.K., Verbakel, H. et al. CaAP2 transcription factor is a candidate gene for a flowering repressor and a candidate for controlling natural variation of flowering time in Capsicum annuum . Theor Appl Genet 128, 1073–1082 (2015). https://doi.org/10.1007/s00122-015-2491-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-015-2491-3