Abstract
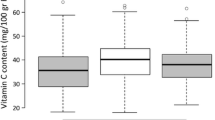
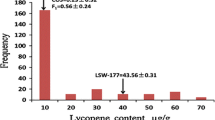
Lycopene content is a key component of tomato (Solanum lycopersicum L.) fruit quality, and is a focus of many tomato-breeding programs. Two QTLs for increased fruit lycopene content, inherited from a high-lycopene S. pimpinellifolium accession, were previously detected on tomato chromosomes 7 and 12 using a S. lycopersicum × S. pimpinellifolium RIL population, and were identified as potential targets for marker-assisted selection and positional cloning. To validate the phenotypic effect of these two QTLs, a BC2 population was developed from a cross between a select RIL and the S. lycopersicum recurrent parent. The BC2 population was field-grown and evaluated for fruit lycopene content using HPLC. Statistical analyses revealed that while lyc7.1 did not significantly increase lycopene content in the heterozygous condition, individuals harboring lyc12.1 in the heterozygous condition contained 70.3 % higher lycopene than the recurrent parent. To eliminate the potential pleiotropic effect of fruit size and minimize the physical size of the lyc12.1 introgression, a marker-assisted backcross program was undertaken and produced a BC3S1 NIL population (n = 1,500) segregating for lyc12.1. Lycopene contents from lyc12.1 homozygous and heterozygous recombinants in this population were measured and lyc12.1 was localized to a 1.5 cM region. Furthermore, we determined that lyc12.1 was delimited to a ~1.5 Mb sequence of tomato chromosome 12, and provided some insight into potential candidate genes in the region. The derived sub-NILs will be useful for transferring of lyc12.1 to other tomato genetic backgrounds and for further fine-mapping and cloning of the QTL.







Similar content being viewed by others
References
Altschul S, Gish W, Miller W, Myers E, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Ashrafi H, Kinkade MP, Foolad MR (2009) A new genetic linkage map of tomato based on a Solanum lycopersicum × S. pimpinellifolium RIL population displaying locations of candidate resistance ESTs. Genome 52:935–956
Ashrafi H, Kinkade M, Merk H, Foolad M (2012) Identification of novel quantitative trait loci for increased lycopene content and other fruit quality traits in a tomato recombinant inbred line population. Mol Breed 30:549–567
Bernacchi D, Beck-Bunn T, Eshed Y, Lopez J, Petiard V, Uhlig J, Zamir D, Tanksley S (1998) Advanced backcross QTL analysis in tomato. I. Identification of QTLs for traits of agronomic importance from Lycopersicon hirsutum. Theor Appl Genet 97:381–397
Brouwer DJ, St. Clair DA (2004) Fine mapping of three quantitative trait loci for late blight resistance in tomato using near isogenic lines (NILs) and sub-NILs. Theor Appl Genet 108:628–638
Brouwer DJ, Jones ES, St. Clair DA (2004) QTL analysis of quantitative resistance to Phytophthora infestans (late blight) in tomato and comparisons with potato. Genome 47:475–492
Cantarel BL, Korf I, Robb SMC, Parra G, Ross E, Moore B, Holt C, Sanchez Alvarado A, Yandell M (2008) MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res 18:188–196
Causse M, Saliba-Colombani V, Lecomte L, Duffé P, Rousselle P, Buret M (2002) QTL analysis of fruit quality in fresh market tomato: a few chromosome regions control the variation of sensory and instrumental traits. J Exp Bot 53:2089–2098
Causse M, Duffe P, Gomez MC, Buret M, Damidaux R, Zamir D, Gur A, Chevalier C, Lemaire-Chamley M, Rothan C (2004) A genetic map of candidate genes and QTLs involved in tomato fruit size and composition. J Exp Bot 55:1671–1685
Chaïb J, Lecomte L, Buret M, Causse M (2006) Stability over genetic backgrounds, generations, and years of quantitative trait locus (QTLs) for organoleptic quality in tomato. Theor Appl Genet 112:934–944
Chen FQ, Foolad MR, Hyman J, St. Clair DA, Beelman RB (1999) Mapping of QTLs for lycopene and other fruit traits in a Lycopersicon esculentum × L. pimpinellifolium cross and comparison of QTLs across tomato species. Mol Breed 5:283–299
Chen Y, Li F, Wurtzel ET (2010) Isolation and characterization of the Z-ISO gene encoding a missing component of carotenoid biosynthesis in plants. Plant Physiol 153:66–79
Doganlar S, Frary A, Ku HM, Tanksley SD (2002) Mapping quantitative trait loci in inbred backcross lines of Lycopersicon pimpinellifolium (LA1589). Genome 45:1189–1202
Eshed Y, Zamir D (1995) An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield-associated QTL. Genetics 141:1147–1162
Foolad MR (2007) Genome mapping and molecular breeding of tomato. Int J Plant Genomics 2007:52 p. Article ID 64358. doi:10.1155/2007/64358:1-52
Frary A, Fulton TM, Zamir D, Tanksley S (2004) Advanced backcross QTL analysis of a Lycopersicon esculentum × L. pennellii cross and identification of possible orthologs in the Solanaceae. Theor Appl Genet 108:485–496
Fraser PD, Bramley PM (2004) The biosynthesis and nutritional uses of carotenoids. Prog Lipid Res 43:228–265
Fulton TM, Beck-Bunn T, Emmatty D, Eshed Y, Lopez J, Petiard V, Uhlig J, Zamir D, Tanksley SD (1997) QTL analysis of an advanced backcross of Lycopersicon peruvianum to the cultivated tomato and comparison with QTLs found in other wild species. Theor Appl Genet 95:881–894
Fulton TM, Grandillo S, Beck-Bunn T, Fridman E, Frampton AJ, Lopez J, Petiard V, Uhlig J, Zamir D, Tanksley SD (2000) Advanced backcross QTL analysis of a Lycopersicon esculentum × Lycopersicon parviflorum cross. Theor Appl Genet 100:1025–1042
Galpaz N, Wang Q, Menda N, Zamir D, Hirschberg J (2008) Abscisic acid deficiency in the tomato mutant high-pigment 3 leading to increased plastid number and higher fruit lycopene content. Plant J 53:717–730
Gardner RG (1988) NC EBR-1 and NC EBR-2 early blight resistant tomato breeding lines. HortScience 23:779–781
Grandillo S, Tanksley SD (1996) QTL analysis of horticultural traits differentiating the cultivated tomato from the closely related species Lycopersicon pimpinellifolium. Theor Appl Genet 92:935–951
Kinkade MP, Foolad MR (2013) Genomics-assisted breeding for tomato fruit quality in the next-generation omics age. In: Varshney RK, Tuberosa R (eds) Genomics assisted plant breeding: yield, quality and abiotic stresses. Wiley-Blackwell Publishers, USA
Lecomte L, Duffé P, Buret M, Servin B, Hospital F, Causse M (2004) Marker-assisted introgression of five QTLs controlling fruit quality traits into three tomato lines revealed interactions between QTLs and genetic backgrounds. Theor Appl Genet 109:658–668
Lewis S, Searle S, Harris N, Gibson M, Iyer V, Richter J, Wiel C, Bayraktaroglu L, Birney E, Crosby M, Kaminker J, Matthews B, Prochnik S, Smith C, Tupy J, Rubin G, Misra S, Mungall C, Clamp M (2002) Apollo: a sequence annotation editor. Genome Biol 33(12):research0082.0081-0082.0014
Liu Y-S, Gur A, Ronen G, Causse M, Damidaux R, Duffe P, Buret M, Hirschberg J, Zamir D (2003) There is more to tomato fruit colour than candidate carotenoid genes. Plant Biotech J 1:195–207
Liu Y, Roof S, Ye Z, Barry C, van Tuinen A, Vrebalov J, Bowler C, Giovannoni J (2004) Manipulation of light signal transduction as a means of modifying fruit nutritional quality in tomato. Proc Natl Acad Sci USA 101:9897–9902
Mueller LA, Solow TH, Taylor N, Skwarecki B, Buels R, Binns J, Lin C-W, Wright MH, Ahrens R, Wang Y, Herbst EV, Keyder ER, Menda N, Zamir D, Tanksley SD (2005a) The SOL genomics network. A comparative resource for Solanaceae biology and beyond. Plant Physiol 138:1310–1317
Mueller LA, Tanksley SD, Giovannoni J, van Eck J, Stack S, Choi D, Kim B, Chen M, Cheng Z, Li C, Ling H, Xue Y, Seymour G, Bishop G, Bryan G, Sharma R, Khurana J, Tyagi A, Chattopadhyay D, Singh N, Stiekema W, Lindhout P, Jesse T, Lankhorst R, Bouzayen M, Shibata D, Tabata S, Granell A, Botella M, Giuliano G, Frusciante L, Causse M, Zamir D (2005b) The tomato sequencing project, the first cornerstone of the international solanaceae project (SOL). Comp Funct Genomics 6:153–158
Neeraja CN, Maghirang-Rodriguez R, Pamplona A, Heuer S, Collard BCY, Septiningsih EM, Vergara G, Sanchez D, Xu K, Ismail AM, Mackill DJ (2007) A marker-assisted backcross approach for developing submergence-tolerant rice cultivars. Theor Appl Genet 115:767–776
Rao AV, Rao LG (2007) Carotenoids and human health. Pharmacol Res 55:207–216
Ronen G, Cohen M, Zamir D, Hirschberg J (1999) Regulation of carotenoid biosynthesis during tomato fruit development: expression of the gene for lycopene epsilon-cyclase is down regulated during ripening and is elevated in the mutant Delta. Plant J 17:341–351
Ronen G, Carmel-Goren L, Zamir D, Hirschberg J (2000) An alternative pathway to beta-carotene formation in plant chloroplast discovered by map-based cloning of Beta and old-gold color mutation in tomato. Proc Natl Acad Sci USA 97:11102–11107
Rousseaux MC, Jones CM, Adams D, Chetelat R, Bennett A, Powell A (2005) QTL analysis of fruit antioxidants in tomato using Lycopersicon pennellii introgression lines. Theor Appl Genet 111:1396–1408
Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386
Sacks EJ, Francis DM (2001) Genetic and environmental variation for tomato flesh color in a population of modern breeding lines. J Am Soc Hort Sci 126:221–226
Saliba-Colombani V, Causse M, Langlois D, Philouze J, Buret M (2001) Genetic analysis of organoleptic quality in fresh market tomato. 1. Mapping QTLs for physical and chemical traits. Theor Appl Genet 102:259–272
Stommel JR, Abbott J, Saftner RA, Camp M (2005) Sensory and objective quality attributes of beta-carotene- and lycopene-rich tomato fruit. J Am Soc Hort Sci 130:244–251
Tanksley SD, Grandillo S, Fulton TM, Zamir D, Eshed Y, Petiard V, Lopez J, Beck-Bunn T (1996) Advanced backcross QTL analysis in a cross between an elite processing line of tomato and its wild relative L. pimpinellifolium. Theor Appl Genet 92:213–224
TFGD (2010) Tomato Functional Genomics Database (http://ted.bti.cornell.edu)
Torres CA, Andrews PK (2006) Developmental changes in antioxidant metabolites, enzymes, and pigments in fruit exocarp of four tomato (Lycopersicon esculentum Mill.) genotypes: β-carotene, high pigment-1, ripening inhibitor, and ‘Rutgers’. Plant Phys Biochem 44:806–818
Wang D, Shi J, Carlson SR, Cregan PB, Ward RW, Diers BW (2003) A low-cost, high-throughput polyacrylamide gel electrophoresis system for genotyping with microsatellite DNA markers. Crop Sci 43:1828–1832
Wu K, Erdman JW, Schwartz SJ, Platz EA, Leitzmann M, Clinton SK, DeGroff V, Willett WC, Giovannucci E (2004) Plasma and dietary carotenoids, and the risk of prostate cancer. Cancer Epidemiol Biomark Prev 13:260–269
Zeng ZB (1994) Precision mapping of quantitative trait loci. Genetics 136:1457–1468
Acknowledgments
This research was supported in part by Agricultural Research Funds administered by the Pennsylvania Department of Agriculture, the Pennsylvania Vegetable Marketing and Research Program, and the College of Agricultural Sciences at the Pennsylvania State University. The authors graciously thank Dr. Randolph Gardner for providing seed of breeding line NCEBR-1, Dr. Hamid Ashrafi for assistance with constructing genetic maps, and all Penn State staff and undergraduates who helped with field experiments and data collection.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by R. Visser.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kinkade, M.P., Foolad, M.R. Validation and fine mapping of lyc12.1, a QTL for increased tomato fruit lycopene content. Theor Appl Genet 126, 2163–2175 (2013). https://doi.org/10.1007/s00122-013-2126-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-013-2126-5