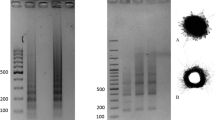
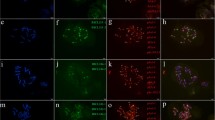
Abstract
Non-denaturing FISH (ND-FISH) was used to compare the distribution of four simple sequence repeats (SSRs)—(AG) n , (AAG) n , (ACT) n and (ATC) n —in somatic root tip metaphase spreads of 12 barley (H. vulgare ssp. vulgare) cultivars, seven lines of their wild progenitor H. vulgare ssp. spontaneum, and four lines of their close relative H. bulbosum, to determine whether the range of molecular diversity shown by these highly polymorphic sequences is reflected at the chromosome level. In both, the cultivated and wild barleys, clusters of AG and ATC repeats were invariant. In contrast, clusters of AAG and ACT showed polymorphism. Karyotypes were prepared after the identification of their seven pairs of homologous chromosomes. Variation between these homologues was only observed in one wild accession that showed the segregation of a reciprocal translocation involving chromosomes 5H and 7H. The two subspecies of H. vulgare analysed were no different in terms of their SSRs. Only AAG repeats were found clustered strongly on the chromosomes of all lines of H. bulbosum examined. Wide variation was seen between homologous chromosomes within and across these lines. These results are the first to provide insight into the cytogenetic diversity of SSRs in barley and its closest relatives. Differences in the abundance and distribution of each SSR analysed, between H. vulgare and H. bulbosum, suggest that these species do not share the same H genome, and support the idea that these species are not very closely related. Southern blotting experiments revealed the complex organization of these SSRs, supporting the findings made with ND-FISH.






Similar content being viewed by others
References
Albert PS, Gao Z, Danilova TV, Birchler JA (2010) Diversity of chromosomal karyotypes in maize and its relatives. Cytogenet Genome Res 129:6–16
Badr A, Müller K, Schäfer-Pregl R, El Rabey H, Effgen S, Ibrahim HH, Pozzi C, Rohde W, Salamini F (2000) On the origin and domestication history of barley (Hordeum vulgare). Mol Biol Evol 17(4):499–510
Blattner FR (2009) Progress in phylogenetic analysis and a new infrageneric classification of the barley genus Hordeum (Poaceae: Triticeae). Breed Sci 59:471–480
Casas AM, Igartua E, Valles MP, Molina-Cano JL (1998) Genetic diversity of barley cultivars grown in Spain, estimated by RFLP, similarity and coancestry coefficients. Plant Breed 117:425–429
Cuadrado A, Jouve N (2002) Evolutionary trends of different repetitive DNA sequences during speciation in genus Secale. J Hered 93:339–345
Cuadrado A, Jouve N (2007a) Similarities in the chromosomal distribution of AG and AC repeats within and between Drosophila, human and barley chromosomes. Cytogenet Genome Res 119:91–9917
Cuadrado A, Jouve N (2007b) The non-random distribution of long clusters of all possible classes of tri-nucleotide repeats in barley chromosomes. Chromosom Res 15:711–720
Cuadrado N, Jouve N (2010) Chromosomal detection of simple sequence repeats (SSRs) using nondenaturing FISH (ND-FISH). Chromosoma 19:495–503
Cuadrado A, Jouve N (2011) Novel simple sequence repeats (SSRs) detected by ND-FISH in heterochromatin of Drosophila melanogaster. BMC Genomics 12:205
Cuadrado A, Schwarzacher T (1998) The chromosomal organization of simple sequence repeats in wheat and rye genomes. Chromosoma 107:587–594
Cuadrado A, Cardoso M, Jouve N (2008) Physical organisation of simple sequence repeats (SSRs) in Triticeae: structural, functional and evolutionary implications. Cytogenet Genome Res 120:210–219
de Bustos A, Cuadrado A, Soler C, Jouve N (1996) Physical mapping of repetitive DNA sequences and 5S and 18S–26S rDNA in five wild species of the genus Hordeum. Chromosom Res 4:491–499
Friebe B, Gill B (1994) C-band polymorphism and structural rearrangements detected in common wheat (Triticum aestivum). Euphytica 78:1–5
Houben A, Pickering R (2009) Applying cytogenetics and genomics to wide hybridisations in the genus Hordeum. In: Feuillet C, Muehlbauer GJ (eds) Genetics and genomics of the Triticeae, plant genetics and genomics: crops and models, vol 7(5). Springer Science, New York, p 137
Kato A (2011) High-density fluorescence in situ hybridization signal detection on barley (Hordeum vulgare L.) chromosomes with improved probe screening and reprobing procedures. Genome 54(2):151–159
Linde-Laursen I, von Botmer R, Jacobsen N (1992) Relationships in the genus Hordeum: Giemsa C-banded karyotypes. Hereditas 116:111–116
Linde-Laursen I, Heslop-Harrison JS, Shepherd KW, Taketa S (1997) The barley genome and its relationship with the wheat genomes. A survey with and internationally agreed recommendation for barley chromosome nomenclature. Hereditas 123:1–16
Morgante M, Oliveri AM (1993) PCR-amplified microsatellites as markers in plant genetics. Plant J 3(1):175–182
Nanda I, Zischler H, Epplen C, Guttenbach M, Schmid M (1991) Chromosomal organization of simple repeated DNA sequences used for DNA fingerprinting. Electrophoresis 12:193–203
Pedersen C, Langridge P (1997) Identification of the entire chromosome complement of bread wheat by two-colour FISH. Genome 40:589–593
Pedersen C, Rasmussen SK, Linde-Laursen I (1996) Genome and chromosome identification in cultivated barley and related species of the Triticeae (Poaceae) by in situ hybridization with the GAA-satellite sequence. Genome 39:93–104
Pickering R, Klatte S, Butler RC (2006) Identification of all chromosome arms and their involvement in meiotic homoeologous associations at metaphase I in 2 Hordeum vulgare L. × Hordeum bulbosum L. hybrids. Genome 49:73–78
Plohl M, Luchetti A, Mestrovic N, Mantovani B (2002) Satellite DNAs between selfishness and functionality: structure, genomics and evolution of tandem repeats in centromeric (hetero)chromatin. Gene 409:72–82
Ramsay L, Macaulay M, Cardle L, Morgante M, Ivanissewith S, Maestri E, Powell W, Waugh R (1999) Intimate association of microsatellite repeats with retrotransposons and other dispersed repetitive elements in barley. Plant J 17:415–525
Röder MS, Korzum V, Wendehake K, Plaschke J, Tixier MH, Leroy P, Ganal MW (1998) A microsatellite map of wheat. Genetics 149(4):2007–2023
Saghai-Maroof MA, Biyashev RM, Yang GP, Zhang Q, Allard RW (1994) Extraordinarily polymorphic microsatellite DNA in barley: species diversity, chromosomal locations, and population dynamics. Proc Natl Acad Sci USA 91:5466–5547
Sanel M, Pickering R, Kumke K, Nasuda S, Houben A (2011) Loss of centromeric histone H3 (CENH3) from centromeres precedes uniparental chromosome elimination in interspecific barley hybrids. Proc Natl Acad Sci USA 108(33):498–505
Schneider A, Linc G, Molnár-Láng M, Graner A (2003) Fluorescence in situ hybridization polymorphism using two repetitive DNA clones in different cultivars of wheat. Plant Breed 122:396–400
Schulte D, Ariyadasa R, Shi B, Fleury D, Saski C, Atkins M, deJong P, Wu CC, Graner A, Langridge P, Stein N (2011) BAC library resources for map-based cloning and physical map construction in barley (Hordeum vulgare L.). BMC Genomics 12:247
Shcherban AB, Vershinin AV (1992) The stretched BamHI-fragment of barley genome containing richly repetitive DNA sequences. Genetika 28:15–21
Svitashev Bryngelsson T, Vershinin A, Pederse C, Sáll T, von Bothmer R (1994) Phylogenetic analysis of the genus Hordeum using repetitive DNA sequences. Theor Appl Genet 89:801–810
Taketa S, Linde-Laursen I, Künzel G (2009) Cytogenetic diversity. In: von Bothmer R, Van Hintum Th, Knüpffer H, Sato D (eds) Diversity in Barley (Hordeum vulgare), pp 97–119. Elsevier Science, Amsterdam
Tautz D, Renz M (1984) Simple sequences are ubiquitous repetitive components of eukaryotic genomes. Nucleic Acids Res 12:4127–4138
Terasawa Y, Rahman SM, Takata K, Ikeda TM (2012) Distribution of Hordoindoline genes in the genus Hordeum. Theor Apple Genet. doi:10.1007/s00122-011-1693-6
von Bothmer R, Jacobsen N (1985) Origin, taxonomy, and related species. In: Rasmusso D (ed) Barley. Agronomy Monographs 26. American Society of Agronomy, Madison, pp 19–56
Xu J, Procunier D, Kasha KJ (1999) Species-specific in situ hybridization of Hordeum bulbosum chromosomes. Genome 33:628–634
Acknowledgments
This study was supported by a grant from the Spanish Ministry of Education and Science (AGL2006-09018-C02). The authors thank Adrian Burton for linguistic assistance.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by B. Friebe.
Electronic supplementary material
Below is the link to the electronic supplementary material.

122_2012_2028_MOESM1_ESM.jpg
Fig. S1 Photomicrographs showing the chromosomal distribution of (AG)10 by ND-FISH in metaphases of the indicated cultivars of H. vulgare ssp. vulgare (a–i) and lines of H. vulgare ssp. spontaneum (m–s). Scale bar = 10 µm (JPEG 3354 kb)

122_2012_2028_MOESM2_ESM.jpg
Fig. S2 Photomicrographs showing the chromosomal distribution of (AAG)5 by ND-FISH in metaphases of the indicated cultivars of H. vulgare ssp. vulgare (a–i) and lines of H. vulgare ssp. spontaneum (m–s). Scale bar = 10 µm (JPEG 3797 kb)

122_2012_2028_MOESM3_ESM.jpg
Fig. S3 Photomicrographs showing the chromosomal distribution of (ACT)5 by ND-FISH in metaphases of the indicated cultivars of H. vulgare ssp. vulgare (a–i) and lines of H. vulgare ssp. spontaneum (m–s). Scale bar = 10 µm (JPEG 4499 kb)

122_2012_2028_MOESM4_ESM.jpg
Fig. S4 Photomicrographs showing the chromosomal distribution of (ATC)5 by ND-FISH in metaphases of the indicated cultivars of H. vulgare ssp. vulgare (a–i) and lines of H. vulgare ssp. spontaneum (m–s). Scale bar = 10 µm (JPEG 4021 kb)
Rights and permissions
About this article
Cite this article
Carmona, A., Friero, E., de Bustos, A. et al. Cytogenetic diversity of SSR motifs within and between Hordeum species carrying the H genome: H. vulgare L. and H. bulbosum L.. Theor Appl Genet 126, 949–961 (2013). https://doi.org/10.1007/s00122-012-2028-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-012-2028-y