Abstract
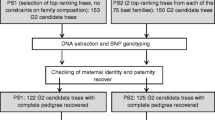
The conventional way to drive modifications in old forest tree seed orchards is to establish progeny trials involving each parent tree and then evaluate its contribution to the performance of the progeny by estimating its general and specific combining ability (GCA and SCA). In this work, we successfully applied an alternative parent selection tactic based on paternity testing of superior offspring derived from a hybrid seed orchard established with a single Eucalyptus grandis seed parents and six E. urophylla pollen parents. A battery of 14 microsatellite markers was used to carry out parentage tests of 256 progeny individuals including two independent samples of selected trees and one control unselected sample, all derived from 6-year-old forest stands in eastern Brazil. Paternity determination was carried out for all progeny individuals by a sequential paternity exclusion procedure. Exclusion was declared only when the obligatory paternal allele in the progeny tree was not present in the alleged parent tree for at least four independent markers to avoid false exclusions due to mutation or null alleles. After maternity checks to identify seed mixtures and selfed individuals, the paternity tests revealed that approximately 29% of the offspring was sired by pollen parents outside the orchard. No selfed progeny were found in the selected samples. Three pollen parents were found to have sired essentially all of the offspring in the samples of selected and non-selected progeny individuals. One of these three parents sired significantly more selected progeny than unselected ones (P≤0.0002 in a Fisher exact test). Based on these results, low-reproductive-successful parents were culled from the orchard, and management procedures were adopted to minimize external pollen contamination. A significant difference (P<0.01) in mean annual increment was observed between forest stands produced with seed from the orchard before and after selection of parents and revitalization of the orchard. An average realized gain of 24.3% in volume growth was obtained from the selection of parents as measured in forest stands at age 2–4 years. The marker-assisted tree-breeding tactic presented herein efficiently identified top parents in a seed orchard and resulted in an improved seed variety. It should be applicable for rapidly improving the output quality of seed orchards, especially when an emergency demand for improved seed is faced by the breeder.
Similar content being viewed by others
References
Bassam BJ, Caetano-Anolles G, Gresshoff PM (1991) Fast and sensitive silver staining of DNA in polyacrylamide gels. Ann Biochem 196:80–83
Bertolucci FL, Resende GD, Penchel R (1995) Produção e utilização de híbridos de eucalipto. Silvicultura 51:12–16
Brandão LG, Campinhos E, Ikemori YK (1984) Brazil’s new forest soars to success. Pulp Pap Int 26:38–40
Brinkmann B, Klintschar M, Neuhuber F, Huhne J, Rolf B (1999) Mutation rate in human microsatellites: influence of the structure and length of the tandem repeat. Am J Hum Genet. 64:1473–1474
Brondani RPV, Brondani C, Tarchini R, Grattapaglia D (1998) Development and mapping of microsatellite based markers in Eucalyptus. Theor Appl Genet 97:816–827
Brondani, RPV, Brondani C, Grattapaglia D (2002) Towards a genus-wide reference linkage map for Eucalyptus based exclusively on highly informative microsatellite markers, Mol Gen Genomics 267:338–347
Brune A, Zobel B. (1981) Genetic base populations, gene pools and breeding populations for Eucalyptus in Brazil. Silvae Genet 30:146–149
Burczyk J, Adams WT, Moran GF, Griffin AR (2002) Complex patterns of mating revealed in a Eucalyptus regnans seed orchard using allozyme markers and the neighbourhood model. Mol Ecol 11:2379–91
Campinhos E (1980) More wood of better quality through intensive silviculture with rapid growth improved Brazilian Eucalyptus. Tappi 63:145–147
Campinhos EN, Peters-Robinson I, Bertolucci FL, Alfenas AC (1998) Interspecific hybridization and inbreeding effect in seed from a Eucalyptus grandis x Eucalyptus urophylla clonal orchard in Brazil. Gen Mol Biol 21:369–374
Chaix G, Gerber S, Razafimaharo V, Vigneron P, Verhaegen D, Hamon S (2003) Gene flow estimation with microsatellites in a Malagasy seed orchard of Eucalyptus grandis. Theor Appl Genet 107:705–712
Eldridge KG, Davidson J, Hardwood C, Van Wyk G (1993) Eucalypt domestication and breeding. Oxford Science Publ, Oxford University Press, Oxford
El-Kassaby YA, Ritland K (1986) The relation of outcrossing and contamination to reproductive phenology and supplemental mass pollination in a Douglas-fir orchard. Silvae Genet 35:240–244
FAO (2000) Global forest resources assessment 2000—main report.FAO Forestry paper http://www.fao.org/forestry/fo/fra/main/index.jsp
Fisher RA (1934) Statistical methods for research workers, 5th edn. Oliver and Boyd, Edinburgh
Gaiotto FA, Bramucci M, Grattapaglia D (1997) Estimation of outcrossing rate in a breeding population of Eucalyptus urophylla s.t. Blake with dominant RAPD and AFLP markers. Theor Appl Genet 95:842–849
Glowatzki-Mullis ML, Gaillard C, Wigger G, Fries R (1995) Microsatellite-based parentage control in cattle. Anim Genet 26:7–12
Grattapaglia D (2000) Molecular breeding of Eucalyptus—state of the art, operational applications and technical challenges. In: Jain SM, Minocha SM (eds) Forestry sciences, vol 64. Molecular biology of woody plants. Kluwer, Dordrecht, pp 451–474
Grattapaglia D, Sederoff R (1994) Genetic linkage maps of Eucalyptus grandis and E. urophylla using a pseudo-testcross mapping strategy and RAPD markers. Genetics 137:1121–1137
Griffin AR, Cotteril PP (1988) Genetic variation in growth of outcrossed, selfed and open-pollinated progenies of Eucalyptus regnans and some implications for breeding strategy. Silvae Genet 37:124–131
Gunn PR, Trueman K, Stapleton P, Klarkowski DB (1997) DNA analysis in disputed parentage: the occurrence of apparently false exclusions of paternity, both at STR loci, in the one child. Electrophoresis 18:1650–1652
Hammond HA, Jin L, Zhong Y, Caskey CT, Chakraborty R (1994) Evaluation of 13 short tandem repeat loci for use in personal identification applications. Am J Hum Genet 55:175–189
Hardner CM, Potts BM (1995) Inbreeding depression and changes in variation after selfing Eucalyptus globulus. Silvae Genet 44:46–54
Harju AM, Nikkanen T (1996) Reproductive success of orchard and non-orchard pollens during different stages of pollen shedding in a Scots pine seed orchard. Can J of For Res 26:1096–1102
Lambeth C, Lee B-C, O’Malley D, Wheeler N (2001) Polymix breeding with parental analysis of progeny: an alternative to full-sib breeding and testing. Theor Appl Genet 103:930–943
Marshall TC, Slate J, Kruuk LE, Pemberton JM (1998) Statistical confidence for likelihood-based paternity inference in natural populations. Mol Ecol 7:639–55
Meagher TR (1986) Analysis of paternity within a natural population of Chamaelirium luteum. 1. Identification of most-likely male parents. Am Nat 128:199–215
Moller A, Wiegand P, Brinkmann B (1995) Observation of null alleles apparently due to deletions. Int J Legal Med 108:90–92
Mora AL, Garcia CH (2000) Eucalypt cultivation in Brazil. Brazilian Society of Silviculture, Brazil
Moran G, Bell JC, Griffin AR (1989) Reduction in levels of inbreeding in a seed orchard of Eucalyptus regnans F. Muell, compared with natural populations. Silvae Genet 38:32–36
Moriguchi Y, Taira H, Tsumura Y (2002) Gene flow of seed orchard in Cryptomeria japonica d.don using microsatellite markers. In: Plant Anim Genome 10. San Diego, abstr 562
Namkoong G, Kang HC, Brouard JS (1988) Tree breeding: principles and strategies. Springer, Berlin Heidelberg New York
Pakkanen A, Nikkanen T, Pulkkinen P (2000) Annual variation in pollen contamination and outcrossing in a Picea abies seed orchard. Scand J For Res 15:399–404
Rhoads DD, Roufa DJ (1990) Seqaid II 3.80. Molecular Genetics Laboratory, Kansas State University, Manhattan, Kan.
Ribeiro VJ, Brondani RPV, Ciampi AY, Grattapaglia D (1998) Paternity determination of superior open pollinated progenies of Eucalyptus based on microsatellite markers [abstr. J24]. Genet Mol Biol 21:212
Valdes AM, Slatkin M, Freimer NB (1993) Allele frequencies at microsatellite loci: the stepwise mutation model revisited. Genetics 133:737–749
Vigneron P (1991) Creation et amelioration de varietes d’hybrides d’Eucalyptus au Congo. In: Intensive forestry: the role of Eucalyptus. Proc IUFRO Symp. Durban, South Africa, pp 345–360
Vigouroux Y, Jaqueth JS, Matsuoka Y, Smith OS, Beavis WD, Smith JS, Doebley J (2002) Rate and pattern of mutation at microsatellite loci in maize. Mol Biol Evol 19:1251–1260
Acknowledgements
We would like to acknowledge Rosana P.V. Brondani for her valuable advice in the genotyping phase. This work was supported by the Brazilian Ministry of Science and Technology—PADCT—FINEP Competitive Grants in Biotechnology to D.G.; Ministry of Education CAPES with a Master fellowship to V.J.R. and the Brazilian National Research Council-CNPq with a research fellowship to D.G.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by D.B. Neale
Rights and permissions
About this article
Cite this article
Grattapaglia, D., Ribeiro, V.J. & Rezende, G.D.S.P. Retrospective selection of elite parent trees using paternity testing with microsatellite markers: an alternative short term breeding tactic for Eucalyptus . Theor Appl Genet 109, 192–199 (2004). https://doi.org/10.1007/s00122-004-1617-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-004-1617-9