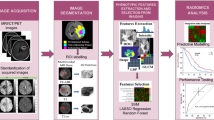
Abstract
Background
Magnetic resonance imaging (MRI) and amino acid positron-emission tomography (PET) of the brain contain a vast amount of structural and functional information that can be analyzed by machine learning algorithms and radiomics for the use of radiotherapy in patients with malignant brain tumors.
Methods
This study is based on comprehensive literature research on machine learning and radiomics analyses in neuroimaging and their potential application for radiotherapy in patients with malignant glioma or brain metastases.
Results
Feature-based radiomics and deep learning-based machine learning methods can be used to improve brain tumor diagnostics and automate various steps of radiotherapy planning. In glioma patients, important applications are the determination of WHO grade and molecular markers for integrated diagnosis in patients not eligible for biopsy or resection, automatic image segmentation for target volume planning, prediction of the location of tumor recurrence, and differentiation of pseudoprogression from actual tumor progression. In patients with brain metastases, radiomics is applied for additional detection of smaller brain metastases, accurate segmentation of multiple larger metastases, prediction of local response after radiosurgery, and differentiation of radiation injury from local brain metastasis relapse. Importantly, high diagnostic accuracies of 80–90% can be achieved by most approaches, despite a large variety in terms of applied imaging techniques and computational methods.
Conclusion
Clinical application of automated image analyses based on radiomics and artificial intelligence has a great potential for improving radiotherapy in patients with malignant brain tumors. However, a common problem associated with these techniques is the large variability and the lack of standardization of the methods applied.
Similar content being viewed by others
Introduction
Neuroimaging is a field of medical imaging that has attracted the most advanced techniques and methods because of the brain’s complex structure and rich endowment with molecular receptors and targets. Most of the available local treatment options, including radiotherapy and neurosurgery, heavily depend on precise knowledge of tumor type, location, and extent that can only be derived from modern imaging methods such as magnetic resonance imaging (MRI) and positron-emission tomography (PET) [1]. The usual sequence of diagnostic and therapeutic procedures in radiotherapy of malignant brain tumors comprises an initial MRI examination, tissue sampling by biopsy or tumor resection, histopathologic determination of tumor type (e.g., glioma vs. metastasis), tumor grade and expression of molecular markers, segmentation and target volume definition, dose planning, decision on concomitant and adjuvant systemic therapy, and, finally, detection and treatment of local relapses or radiation-induced injury of the brain during follow-up.
Each of these steps can be aided by application of radiomics and image-based machine learning. In many instances, the success of these procedures depends on the complexity level of the applied imaging techniques. Conventional MRI techniques are T1 weighted, T2 weighted, and fluid attenuated inversion recovery (FLAIR) images, while advanced MRI methods include perfusion-weighted imaging (PWI), diffusion-weighted imaging (DWI) including its variants such as diffusion-tensor imaging (DTI), and MR spectroscopy (MRS). The applied machine learning methods mainly comprise feature-based radiomics, where modeling is based on a mathematically predefined set of features extracted from a manually segmented image, and deep learning approaches where the complete model is trained on the imaging data and does not necessarily require a segmented input. However, the applied imaging and computational methods vary substantially and are far from being standardized. The following review aims at providing an overview of the results that were achieved by different machine learning approaches for patients with malignant glioma and metastases.
Glioma
Pre-therapeutic classification and molecular characterization
In 2016, the World Health Organization (WHO) published revised guidelines for the classification of tumors of the central nervous system. The most important glioma types in which radiotherapy is integrated into the treatment concept are WHO grade II diffuse astrocytomas and oligodendrogliomas, WHO grade III anaplastic astrocytomas and anaplastic oligodendrogliomas, and WHO grade IV glioblastomas. The classification is based on both tumor histology as well as the presence of a mutation in the isocitrate dehydrogenase (IDH) gene and the loss of heterozygosity (LOH) of the 1p/19q chromosome arms, which allows an integrated diagnosis according to the WHO classification 2016 [2]. Possible treatment strategies for radiotherapy of glioma patients including concomitant and adjuvant chemotherapy are predominantly based on the WHO grade and, more importantly, on molecular characteristics of the tumor [3], namely the IDH genotype [4], the 1p/19q status [5,6,7], and the O6-methylguanine-DNA methyltransferase (MGMT) promoter methylation status [8,9,10]. The diagnosis of a malignant glioma and the determination of the molecular markers are usually based on tissue samples obtained by tumor resection or stereotactic biopsy. However, approximately 15% of glioma are unresectable [11, 12], and stereotactic biopsy carries a measurable risk for morbidity, especially in the older population [13]. Therefore, several studies have investigated the application of radiomics for determination of WHO grade [14,15,16,17,18,19,20,21] and molecular characteristics [22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40] in glioma. Results of selected reports applying radiomics or other machine learning methods on conventional MRI, advanced MRI, and PET are reported in more detail in the following sections.
Determination of WHO grade in patients with newly diagnosed gliomas
Ditmer and colleagues [15] investigated the diagnostic accuracy of histogram-based radiomics analysis of post-contrast T1-weighted MRI for the differentiation of high-grade from low-grade gliomas in 94 patients. The highest diagnostic accuracy was achieved by using the mean of the histograms (area under curve, AUC, 0.9; sensitivity 93%; specificity 86%). However, the dataset was highly unbalanced and parameter combinations were not investigated. Cho and colleagues [14] applied a radiomics approach using different machine learning classifiers for glioma grading based on 285 datasets from the Brain Tumor Segmentation (BraTS) Challenge 2017 [41] comprising pre- and post-contrast T1-weighted, T2-weighted, and FLAIR MRI. After calculation of 468 radiomics features, 5 were selected for use in a random forest classifier that showed the highest AUC of 0.92 after fivefold cross validation.
Besides conventional MRI, several groups have also used advanced MRI methods in combination with radiomics analyses for glioma grading. Sengupta and colleagues [17] used features extracted from conventional MRI and from PWI. The model generated by a support vector machine classifier yielded low classification errors ranging from 3.7% for WHO grade II vs. III up to 9.4% for WHO grade II vs. grade III vs. grade IV. Similarly, Takahashi and colleagues [18] evaluated T2-weighted MRI in combination with DWI for differentiation of glioblastoma from lower-grade glioma in 54 patients. After feature selection, a support vector machine classifier using 6 parameters extracted from DWI demonstrated the best performance in the test dataset (AUC, 0.93). Besides MRI radiomics, Pyka and colleagues evaluated the ability of amino acid PET radiomics using the tracer O‑(2-[18F]fluoroethyl)-L-tyrosine (FET) for glioma grading [40]. The combination of textural features calculated from the grey-level neighborhood difference matrix and the metabolic tumor volume yielded a diagnostic accuracy of 85% for the differentiation of WHO grade III and IV gliomas.
Yang and colleagues [21] investigated the usefulness of deep learning-based radiomics using convolutional neural networks (CNNs) for glioma grading in a group of 113 patients. Conventional and advanced MRI including PWI and DWI were available and two commonly used CNNs (AlexNet and GoogLeNet) were explored. The CNNs were applied in two forms. First, only the network structure was taken over and the networks were trained from scratch using the MR images. Second, the networks were used with the connection weights already pretrained on a dataset of 1.2 million non-medical images from “ImageNet,” a large-scale natural image database, and were only finetuned by retraining the first convolutional layer and the final fully connected layers on the MR images. The pretrained, finetuned GoogLeNet showed the best classification accuracy in the test dataset (AUC 0.94). This study demonstrates that pretrained CNNs can be useful for clinical decision-making in patients with gliomas, especially when the number of available patients is low. In summary, assessment of WHO grade in newly diagnosed gliomas by means of machine learning methods achieves an accuracy of approximately 90% and may thus be of clinical benefit in patients unsuitable for resection or biopsy.
Determination of IDH genotype and 1p/19q status in patients with newly diagnosed gliomas
Once the diagnosis of a diffuse glioma has been made, usually from tissue sampling, the treatment plan, including dose, type, and fractionation of radiotherapy as well as the sequence of chemotherapy, is mainly determined by the molecular characteristics of the tumor based on the WHO classification [3]. Many groups have shown that these may also be derived non-invasively from imaging by application of machine learning [22,23,24,25, 33,34,35, 38, 39, 42, 43]. Zhang and co-workers [43] used conventional MRI as well as DWI and extracted a total of 2970 features from 120 patients with WHO grade III and IV gliomas. Finally, a subset of 386 features was used to build a model based on a random forest classification, resulting in an accuracy of 89% for IDH prediction in the validation dataset. In WHO grade II and III gliomas, Zhou and colleagues [39] showed that a logistic regression model with only three features obtained from conventional MRI of 165 patients predicted the IDH genotype as well as the 1p/19q co-deletion status with AUC values of 0.86 and 0.96, respectively. In a subsequent study [38], Zhou and colleagues extracted radiomics features from conventional MRI of a large multicentric dataset of more than 500 patients. The model was built using a random forest classifier and validated using another set of MR images from The Cancer Imaging Archive (TCIA). The IDH genotype could be predicted with an AUC of 0.92 in the test cohort. Lu and colleagues [34] used conventional MRI data from 214 glioma patients from TCIA. Additionally, 70 patients with MRI scans from different institutions were used for model testing. A multi-level machine learning model based on support vector machine classifiers was used and allowed classification of IDH and 1p/19q status with accuracies of 88% and 96%, respectively. Lohmann and colleagues [33] used FET PET radiomics for preoperative prediction of IDH genotype in a cohort of 84 glioma patients and identified a simple two-parameter logistic regression model that achieved a diagnostic accuracy of 80% after 10-fold cross validation. A subgroup analysis of 28 patients measured on a high-resolution BrainPET scanner showed the highest accuracy of 86% after 10-fold cross validation.
Again, deep learning-based radiomics approaches have also been investigated for prediction of molecular characteristics in gliomas. Chang and co-workers [22] used a residual CNN for IDH genotype prediction on conventional MRI from a large cohort of 496 glioma patients from three different institutions and achieved an accuracy of 86% in an independent test set. The accuracy could be further increased to 89% by the incorporation of age at diagnosis. Eichinger and colleagues [24] used a set of local binary pattern features extracted from T2-weighted MRI and DTI for training of a CNN with a single hidden layer. The prediction of IDH genotype yielded a diagnostic accuracy of 95% in the test dataset. These results suggest that radiomics has the potential to substitute neuropathological assessment of IDH mutation and 1p/19q LOH status in glioma patients in whom tissue sampling is not feasible.
Determination of MGMT promoter methylation status
The methylation status of the MGMT promoter is of great value for predicting the response to alkylating chemotherapy and has also been determined using advanced image analyses. Korfiatis and colleagues [28] predicted the MGMT promoter methylation status in patients with glioblastoma with a sensitivity of 80% and a specificity of 81% using a four-parameter model based on T2-weighted MRI. Xi and co-workers [36] calculated 1665 radiomics features from conventional MRI and generated a model from a subset of 36 features, resulting in an accuracy of 87% in the validation data and 80% in a test dataset. Similarly, Li and colleagues [31] extracted 1705 radiomics features from conventional MRI and built a predictive random forest model using a subset of six features. The final model resulted in an AUC of 0.88. The combination of clinical factors with radiomic features did not further improve model performance. Kong and colleagues [27] used FDG PET-based radiomics in 107 patients with primary glioma and extracted a total of 1561 features for prediction of MGMT promoter methylation status. Five radiomics features were finally selected to construct the radiomics signature using a support vector machine classifier. The model achieved an AUC of 0.94 in the validation and 0.86 in the test cohort.
By means of a bidirectional, recurrent CNN that leverages the spatial aspects of three-dimensional MRI scans, Han and Kamdar [25] obtained an accuracy of 67% in the validation dataset and 62% in the test data. Korfiatis and colleagues [29] compared three different residual deep neural network (ResNet) architectures for the prediction of MGMT promoter methylation status based on conventional MRI data from 155 patients. The authors found that the ResNet50 with a 50-layer architecture was the best-performing model, achieving an accuracy of 95% in the test dataset. Thus, promising results also exist regarding non-invasive, image-based assessment of MGMT promoter methylation status.
Image segmentation and delineation of planning target volumes
Malignant gliomas tend to grow in typical patterns and induce a number of characteristic tissue changes. The main tumor compartments, also called “segments” in image processing, comprise the necrotic core, contrast-enhancing tumor, non-enhancing tumor, and perifocal edema. Fast and reliable labeling (contouring) of these segments is a crucial task in many areas of neuro-oncology such as radiotherapy and image-based follow-up [44]. Numerous algorithms have been developed for this purpose [45] and a periodically held challenge (the Multimodal Brain Tumor Image Segmentation Benchmark, BRATS) has been set up to compare their efficacy [41, 46]. Today, the best-performing tools are usually based on CNNs [47, 48], which achieve high segmentation accuracies (Dice similarity coefficients) where almost 90% of the voxels are correctly labeled, which is the order of magnitude that experienced physicians can achieve [44]. In radiotherapy planning for malignant glioma, delineation of gross tumor volume (GTV) and clinical target volume (CTV) could in principle be automatically performed by these software tools [49]. A basic requirement is that segmentation should not only work for untreated tumors but also in less well-defined situations, e.g., after tumor resection, which is the most common situation in radiotherapy for malignant glioma. Vendors of radiotherapy planning systems should aim at integration of these tools into their software in order to accelerate and standardize the contouring process. A powerful segmentation toolkit for scientific use is publicly available at: https://github.com/neuronflow/BraTS-Toolkit.
Prediction of local relapse location after radiotherapy
In malignant glioma, approximately 80–90% of recurrences that develop after macroscopic tumor resection and postoperative local irradiation with doses of approximately 60 Gy are located within a distance of 2 cm to the margin of the resection cavity and will occur within 6–9 months after initiation of radiotherapy [50,51,52,53,54]. This observation was used for recommending the size of the geometrical margin between the CTV and the GTV in both European and North American guidelines [55, 56]. However, this procedure still results in considerably large volumes of irradiated brain, and almost all attempts to escalate the radiation dose up to 80–90 Gy within a geometrically defined CTV have failed to improve the prognosis of these patients [57,58,59,60]. Assuming that the direction of tumor spread is foreseeable and that a dose–response relationship exists for malignant glioma, prediction of the precise location of a recurrence would be of significant value. Ideally, a so-called tumor infiltration map of the peritumoral region should be generated that depicts the areas with the highest risk of recurrence, which, in turn, could be targeted by higher doses. Several approaches including conventional imaging analysis methods and radiomics have been applied to achieve this goal.
Amino acid PET was used for definition of the CTV in a small study with focal dose escalation to 72 Gy. Although the results were encouraging in terms of survival [61], there was only a small overlap between the PET signals at recurrence and the PET signals used for dose planning [62]. In a comparable study, 63% of the recurrent tumor volume was located outside the PET-defined GTV [63]. However, it was shown that the GTV–CTV margins could be reduced by 4 mm for the PET-defined GTV in comparison to the MRI-defined GTV in order to include 100% of the recurrences.
In addition to PET, advanced MR imaging techniques have been applied for predicting the spread of glioma cells. By use of DWI and DTI, regions with restricted diffusion due to increased cellular density can be identified. Indeed, fractional anisotropy (FA), a measure of directed diffusion, was found to be significantly lower in regions with later tumor recurrence [64]. Also, areas of the preoperative peritumoral region with a lowered apparent diffusion coefficient (ADC) overlapped with the region of the later recurrences by 60% [65]. DTI and further advanced MR can also be used to depict the orientation and density of white-matter tracts in the vicinity of the macroscopic tumor. By this approach, it was shown that tumor growth follows the tracts to some extent [66] and that this fact can be used to foresee which brain regions are predominantly affected by recurrent tumor growth [67].
However, the most promising approaches are probably those that use advanced, multiparametric MR imaging in conjunction with radiomics and machine learning. In a series of investigations, a group from the University of Philadelphia applied conventional and advanced MR techniques (pre- and post-contrast T1-weighted MRI, T2-weighted MRI, PWI, and DTI) to determine FA, radial diffusivity, axial diffusivity, ADC, relative cerebral blood volume, and a number of associated first-and second-order features in each voxel of the peritumoral region. A model was trained to predict the voxel’s risk for being involved in a recurrent tumor, which resulted in a tumor infiltration map that had an overall accuracy of approximately 90% [68,69,70]. If these maps were routinely available, postoperative radiotherapy of malignant glioma could be far more individualized compared to the present practice.
Prediction of progression-free and overall survival
In radiotherapy planning, not only the putative location of a future recurrence, but also the time interval to progression is of major importance. Methods including radiomics and machine learning have therefore also been used to predict progression-free (PFS) and overall survival (OS), which are both closely related to the time to progression. When using radiomic features from standard preoperative MRI of the primary tumor or the postoperative peritumoral region, predictive models for PFS and OS were developed that outperformed those based on clinical features alone [71, 72]. Also, the performance of these models improved significantly when using advanced MR imaging features, e.g., from PWI or DTI, and combining them with clinical factors [73,74,75,76]. In the era of prognostic and predictive molecular markers, the imaging features may seem of less importance. However, Kickingereder and colleagues demonstrated in a cohort of IDH-wildtype glioblastoma patients with known MGMT methylation status that the addition of radiomic features to a comprehensive model comprising clinical, therapeutic, and molecular features could still improve the prediction accuracy [74].
Differentiation of tumor progression from pseudoprogression
The term pseudoprogression describes the occurrence of a progressive enhancing lesion on MRI within 12 weeks after radiotherapy alone or radiotherapy with concomitant and adjuvant temozolomide chemotherapy in patients with malignant gliomas with spontaneous improvement without any treatment change [77, 78]. Due to its clinical importance, this time-dependent definition of pseudoprogression was included in the recommendations of the Response Assessment in Neuro-Oncology (RANO) working group [79]. Several studies have already demonstrated the usefulness of amino acid PET as well as PWI for this challenging clinical task [77, 80,81,82]. However, radiomics and machine learning might add important diagnostic information to further improve the diagnostic performance.
Hu and colleagues [83] used conventional and advanced MRI including DWI and PWI for differentiation of tumor progression from pseudoprogression in 31 patients who underwent radiochemotherapy after surgical resection. An eight-dimensional feature vector was constructed, and a support vector machine classifier yielded an AUC of 0.94 (sensitivity 90%; specificity 94%). These findings were confirmed by the study from Kim and colleagues [84]. Therein, a multiparametric model incorporating conventional MRI in combination with DWI and PWI showed a significantly better performance than other models based on single imaging contrasts with an AUC of 0.85 in the test dataset.
FET PET radiomics has also been investigated for differentiation of tumor progression from pseudoprogression. Kebir and co-workers [85] used FET PET scans of 14 patients and applied an unsupervised consensus clustering algorithm for the diagnosis of pseudoprogression, resulting in a diagnostic accuracy of 75%. However, the number of patients was very low and the model was not further validated. Lohmann and colleagues [86] also investigated the potential of FET PET radiomics for this purpose. Thirty-five glioblastoma patients with imaging findings suspicious for pseudoprogression within 12 weeks after completion of radiochemotherapy were included. The final logistic regression model used three textural features and yielded a diagnostic accuracy of 92% after 10-fold cross validation in the training and 86% in the test dataset. Again, the model has to prove its performance in a larger cohort study.
A few studies have applied deep learning-based radiomics to classify seemingly progressing lesions. Jang and co-workers [87] used post-contrast MR images from 78 patients from two institutions. The developed CNN incorporated both clinical and imaging features and achieved an AUC of 0.83 in the test dataset. Interestingly, models based solely on clinical or imaging features showed inferior performance, highlighting the importance of incorporation of clinical features. Bachhi and colleagues [88] used a CNN based on conventional MRI and DWI. In a cohort of 55 patients, the CNN model based on DWI and FLAIR sequences in combination achieved the highest diagnostic accuracy of 82% in the test dataset. Akbari and colleagues [89] also used multiparametric MRI data consisting of conventional MRI as well as DWI and PWI in a cohort of 63 patients where detailed histopathological confirmation of the diagnosis was available for all patients. Classical radiomics features as well as deep learning-based features were combined for classification using support vector machines. The final model achieved a diagnostic accuracy for the diagnosis of pseudoprogression of 87% (AUC 0.92) after leave-one-out cross validation. The accuracy in the inter-institutional test cohort was 75% (AUC 0.8). Of note, the workflow can be easily adapted, as the authors used a freely available software toolkit (Cancer Imaging Phenomics Toolkit). Recently, Li and co-workers [90] introduced a novel feature-learning method based on deep convolutional generative adversarial networks (DCGAN) and a CNN (AlexNet), termed DC-AL GAN. Discriminative features identified by DC-AL GAN were used for classification by support vector machines, resulting in a diagnostic accuracy of 92% (AUC, 0.95) after 10-fold cross validation.
Differential diagnosis
Glioblastoma and brain metastases are the two most common malignant brain tumors in adults and often present similar clinical and imaging characteristics on conventional MRI [91, 92]. Consequently, differential diagnosis based solely on clinical presentation and MRI alone is often challenging.
Qian and colleagues [93] addressed this question using MRI radiomics. A large group of 412 patients with untreated brain metastases and treatment-naive, newly diagnosed glioblastoma was divided into a training and a test cohort. Tumors were segmented manually and 1303 radiomic features were calculated on contrast-enhanced MR images. The best classifier that showed a high predictive performance in the test cohort (AUC 0.90) was a support vector machine algorithm that used the least absolute shrinkage and selection operator (LASSO) for feature selection. Also, the classifier showed a better performance than experienced neuroradiologists.
Artzi and colleagues [94] extracted 760 radiomics features from contrast-enhanced MR images of 439 patients with brain metastases or glioblastoma. After image preprocessing and semi-automatic tumor segmentation using a region-growing algorithm, feature selection and model generation was performed. Interestingly, the authors identified the same support vector machine algorithm as the study by Qian and colleagues described above to have the highest predictive performance in the test cohort (AUC 0.96) for the differentiation of brain metastases from glioblastoma.
Brain metastases
Detection and automatic segmentation of brain metastases
Nowadays, the dominant type of radiotherapy for a limited number and limited total volume of brain metastases is radiosurgery [95]. Particularly when multiple metastases are present, manual contouring is a laborious task. Automatic segmentation of brain metastases for use in radiosurgery planning differs in several aspects from that of gliomas. As metastases are thought to have a low infiltrative potential, definition of the GTV is mainly based on the T1-weighted, contrast-enhanced MR images with no or only small GTV–CTV margins, while other tumor segments are usually not considered. However, some metastases leave the blood–brain barrier intact, while others grow adjacent to other contrast-enhancing structures or resemble small vessels in appearance. Therefore, automatic segmentation algorithms should be able to both reliably identify brain metastases (measured by the detection rate and the false-positive rate) and to accurately contour them (measured by the Dice similarity coefficient). To achieve this goal, most groups adapted existing CNN solutions. Liu and colleagues applied a modified version of the widely used DeepMedic CNN for use with small metastases (<1.5 cm) and achieved an average Dice coefficient of 0.67 on contrast-enhanced MR images [96]. The same CNN was used in a dataset were contrast-enhanced and FLAIR MR images were available and achieved a Dice coefficient of 0.79 with a typical detection rate of 93%, which came at the cost of a false-positive rate of 7.8 metastases per patient [97]. A group from Stanford used a CNN based on GoogLeNet on contrast-enhanced and FLAIR MR images and achieved a Dice coefficient of 0.79. However, the sensitivity was only 50% for metastases <7 mm but 100% for lesions >22 mm at an average false-positive rate of 8.3 metastases per patient. In summary, the contouring performance of these automated systems seems to be at the edge of suitability for clinical use, while secure detection of small metastases is at present only possible if a considerable number of falsely labeled non-tumor regions is accepted.
Prediction of local response after radiosurgery of brain metastases
It is widely believed that the main determinants of local response to radiosurgery are tumor volume and prescription dose; however, the response to radiosurgery is still difficult to predict in a subset of brain metastases. This may be partly due to the more radioresistant tumor cells residing at the border of a necrotic core that is present in a proportion of the metastases, and to differences in the tumor vasculature that takes part in promoting the response to single high-dose irradiation [98]. As these tumor properties may be reflected in the radiological appearance of the metastasis and its surrounding area, radiomics is a promising tool for predicting response after radiosurgery. Indeed, simple features such as the presence of a necrotic core [99], the fraction of contrast-enhancing tumor tissue [100], or the extension of the perifocal edema [101] have been shown to impact on response or survival. The experience with more advanced imaging methods and radiomics is limited so far. However, by training a simple CNN on image patches from a cranial computed tomography, Cha and co-workers were able to classify tumors as responding vs. non-responding with an AUC of approximately 0.8 [102]. In a recent report, a classic radiomics approach was applied with features extracted from contrast-enhanced and FLAIR MR images of the core metastasis and its immediate vicinity. By adding any of the top-ranked radiomic features to a model based on pure clinical and dosimetric factors (dose, isodose, diameter, number and location of metastases, previous whole-brain radiotherapy), the AUC for predicting response could be increased from approximately 0.7 to 0.8 [103].
Differentiation of radiation injury from local brain metastasis relapse
Following radiosurgery for brain metastases, radiation injury of the brain tissue may develop in approximately 5–20% of patients [104]. Radiation injury is usually suspected if new contrast-enhancing lesions appear in or adjacent to the GTV and is often indistinguishable from local brain metastasis relapse using conventional MRI alone. The pathological mechanisms involved in these two processes differ substantially [105] and should therefore be distinguishable by a radiomics analysis of the associated image changes.
Peng and colleagues [106] evaluated the usefulness of MRI radiomics for this important question. Sixty-six patients with 82 lesions treated with stereotactic radiosurgery and imaging findings on contrast-enhanced and FLAIR MRI suspicious for tumor recurrence were included in the study. Fifty-one radiomics features (3 shape features, 14 histogram-based features, and 34 textural features) were extracted for each lesion on each MRI contrast. Models were generated using the IsoSVM algorithm, which performs both feature selection and classification [107]. No separate dataset was available for model testing. However, cross validation was performed to assess overall model performance. The model reached an area under the receiver operating characteristic curve (AUC) of 0.81, with a specificity of 65% and a sensitivity of 87%. On the contrary, experienced radiologists could only classify 73% of the cases, with a sensitivity of 97% and a specificity of only 19%.
Similarly, Zhang and colleagues [108] used pre- and post-contrast T1-weighted MR images, T2, and FLAIR from 87 patients after Gamma Knife (Elekta, Stockholm, Sweden) radiosurgery to calculate 285 radiomics features. Interestingly, imaging data from two timepoints were available, so that the authors also investigated feature reproducibility to identify a feature subset with reproducible values. Changes in radiomics features (so-called delta radiomics) from one follow-up timepoint to the next were evaluated and used for differentiation of radiation necrosis and tumor progression. The final model generated by an ensemble classifier had an overall predictive accuracy of 73% and an AUC of 0.73 after cross validation. Again, no separate test dataset was available.
Besides MRI, amino acid PET has also been investigated to evaluate radiomics for the differentiation of treatment-related changes from brain metastasis recurrence. It has been demonstrated that evaluation of the time–activity curves (TAC) that represent the tracer uptake over time is helpful for differentiation of treatment-related changes from brain metastasis recurrence [109]. However, this requires a time-consuming dynamic FET PET scan of at least 40 min acquisition time or more. Therefore, Lohmann and colleagues [110] calculated 62 textural parameters on static FET PET scans from 47 patients with MRI findings suspicious for tumor recurrence after radiosurgery. Combinations of conventional FET PET and textural features were investigated using ROC analysis without prior feature selection. The diagnostic accuracy of conventional FET PET parameters was in the range of 81–83% and could be slightly increased to 85% when combined with textural features. However, no dataset for validation or testing was available. In a subsequent study, Lohmann and colleagues [111] investigated the value of combining FET PET and MRI radiomics for the differentiation of treatment-related changes from brain metastasis recurrence. Fifty-two patients with newly or progressive contrast-enhancing lesions on MRI after radiotherapy were additionally investigated using FET PET. After feature selection, logistic regression models limited to a maximum of five parameters to avoid over-fitting were generated for the combined PET/MRI features and for each modality separately and validated using cross validation; no test dataset was available. The highest diagnostic accuracy of 89% (specificity 96%, sensitivity 85%) was achieved by the combination of MRI and FET PET features, suggesting that the combined FET PET/MRI radiomics analysis encoded more diagnostic information than either modality alone. In summary, both MRI- and PET-based radiomics achieve classification accuracies far above chance for differentiation of radiation injury from tumor recurrence and thus demonstrate the potential of this approach.
Conclusion
Feature-based radiomics and deep learning-based machine learning methods can be used to facilitate and automate many of the diagnostic and therapeutic steps needed in radiotherapy of malignant brain tumors with high accuracy. A common problem associated with the application of the derived models is the large variability and the lack of standardization of both image acquisition and computational methods [112]. From the perspective of radiotherapy, the most promising applications include automatic segmentation of the target volumes, prediction of the location and timepoint of local recurrences of glioma by tumor infiltration maps, and differentiation of treatment-related tissue changes from true recurrences.
References
Langen KJ, Galldiks N, Hattingen E, Shah NJ (2017) Advances in neuro-oncology imaging. Nat Rev Neurol 13:279–289
Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW (2016) The 2016 world health organization classification of tumors of the central nervous system: a summary. Acta Neuropathol 131:803–820
Weller M, van den Bent M, Tonn JC, Stupp R, Preusser M, Cohen-Jonathan-Moyal E, Henriksson R, Rhun EL, Balana C, Chinot O, Bendszus M, Reijneveld JC, Dhermain F, French P, Marosi C, Watts C, Oberg I, Pilkington G, Baumert BG, Taphoorn MJB, Hegi M, Westphal M, Reifenberger G, Soffietti R, Wick W, European Association for Neuro-Oncology (EANO) Task Force on Gliomas (2017) European association for neuro-oncology (EANO) guideline on the diagnosis and treatment of adult astrocytic and oligodendroglial gliomas. Lancet Oncol 18:e315–e329
Hartmann C, Hentschel B, Wick W, Capper D, Felsberg J, Simon M, Westphal M, Schackert G, Meyermann R, Pietsch T, Reifenberger G, Weller M, Loeffler M, von Deimling A (2010) Patients with IDH1 wild type anaplastic astrocytomas exhibit worse prognosis than IDH1-mutated glioblastomas, and IDH1 mutation status accounts for the unfavorable prognostic effect of higher age: implications for classification of gliomas. Acta Neuropathol 120:707–718
Chamberlain MC, Born D (2015) Prognostic significance of relative 1p/19q codeletion in oligodendroglial tumors. J Neurooncol 125:249–251
Speirs CK, Simpson JR, Robinson CG, DeWees TA, Tran DD, Linette G, Chicoine MR, Dacey RG, Rich KM, Dowling JL, Leuthardt EC, Zipfel GJ, Kim AH, Huang J (2015) Impact of 1p/19q codeletion and histology on outcomes of anaplastic gliomas treated with radiation therapy and temozolomide. Int J Radiat Oncol Biol Phys 91:268–276
van den Bent MJ, Brandes AA, Taphoorn MJ, Kros JM, Kouwenhoven MC, Delattre JY, Bernsen HJ, Frenay M, Tijssen CC, Grisold W, Sipos L, Enting RH, French PJ, Dinjens WN, Vecht CJ, Allgeier A, Lacombe D, Gorlia T, Hoang-Xuan K (2013) Adjuvant procarbazine, lomustine, and vincristine chemotherapy in newly diagnosed anaplastic oligodendroglioma: long-term follow-up of EORTC brain tumor group study 26951. J Clin Oncol 31:344–350
Hegi ME, Liu L, Herman JG, Stupp R, Wick W, Weller M, Mehta MP, Gilbert MR (2008) Correlation of O6-methylguanine methyltransferase (MGMT) promoter methylation with clinical outcomes in glioblastoma and clinical strategies to modulate MGMT activity. J Clin Oncol 26:4189–4199
Buckner JC, Shaw EG, Pugh SL, Chakravarti A, Gilbert MR, Barger GR, Coons S, Ricci P, Bullard D, Brown PD, Stelzer K, Brachman D, Suh JH, Schultz CJ, Bahary JP, Fisher BJ, Kim H, Murtha AD, Bell EH, Won M, Mehta MP, Curran WJ Jr. (2016) Radiation plus procarbazine, CCNU, and vincristine in low-grade glioma. N Engl J Med 374:1344–1355
Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani L, Bromberg JE, Hau P, Mirimanoff RO, Cairncross JG, Janzer RC, Stupp R (2005) MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med 352:997–1003
Nieder C, Grosu AL, Astner S, Molls M (2005) Treatment of unresectable glioblastoma multiforme. Anticancer Res 25:4605–4610
Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross JG, Eisenhauer E, Mirimanoff RO, European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups, National Cancer Institute of Canada Clinical Trials Group (2005) Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 352:987–996
Kellermann SG, Hamisch CA, Ruess D, Blau T, Goldbrunner R, Treuer H, Grau SJ, Ruge MI (2017) Stereotactic biopsy in elderly patients: risk assessment and impact on treatment decision. J Neurooncol 134:303–307
Cho HH, Lee SH, Kim J, Park H (2018) Classification of the glioma grading using radiomics analysis. PeerJ 6:e5982
Ditmer A, Zhang B, Shujaat T, Pavlina A, Luibrand N, Gaskill-Shipley M, Vagal A (2018) Diagnostic accuracy of MRI texture analysis for grading gliomas. J Neurooncol 140:583–589
Li-Chun Hsieh K, Chen CY, Lo CM (2017) Quantitative glioma grading using transformed gray-scale invariant textures of MRI. Comput Biol Med 83:102–108
Sengupta A, Ramaniharan AK, Gupta RK, Agarwal S, Singh A (2019) Glioma grading using a machine-learning framework based on optimized features obtained from T1 perfusion MRI and volumes of tumor components. J Magn Reson Imaging. https://doi.org/10.1002/jmri.26704
Takahashi S, Takahashi W, Tanaka S, Haga A, Nakamoto T, Suzuki Y, Mukasa A, Takayanagi S, Kitagawa Y, Hana T, Nejo T, Nomura M, Nakagawa K, Saito N (2019) Radiomics analysis for glioma malignancy evaluation using diffusion kurtosis and tensor imaging. Int J Radiat Oncol Biol Phys. https://doi.org/10.1016/j.ijrobp.2019.07.011
Tian Q, Yan LF, Zhang X, Zhang X, Hu YC, Han Y, Liu ZC, Nan HY, Sun Q, Sun YZ, Yang Y, Yu Y, Zhang J, Hu B, Xiao G, Chen P, Tian S, Xu J, Wang W, Cui GB (2018) Radiomics strategy for glioma grading using texture features from multiparametric MRI. J Magn Reson Imaging 48:1518–1528
Vamvakas A, Williams SC, Theodorou K, Kapsalaki E, Fountas K, Kappas C, Vassiou K, Tsougos I (2019) Imaging biomarker analysis of advanced multiparametric MRI for glioma grading. Phys Med 60:188–198
Yang Y, Yan LF, Zhang X, Han Y, Nan HY, Hu YC, Hu B, Yan SL, Zhang J, Cheng DL, Ge XW, Cui GB, Zhao D, Wang W (2018) Glioma grading on conventional MR images: a deep learning study with transfer learning. Front Neurosci 12:804
Chang K, Bai HX, Zhou H, Su C, Bi WL, Agbodza E, Kavouridis VK, Senders JT, Boaro A, Beers A, Zhang B, Capellini A, Liao W, Shen Q, Li X, Xiao B, Cryan J, Ramkissoon S, Ramkissoon L, Ligon K, Wen PY, Bindra RS, Woo J, Arnaout O, Gerstner ER, Zhang PJ, Rosen BR, Yang L, Huang RY, Kalpathy-Cramer J (2018) Residual convolutional neural network for the determination of IDH status in low- and high-grade gliomas from MR imaging. Clin Cancer Res 24:1073–1081
Chang P, Grinband J, Weinberg BD, Bardis M, Khy M, Cadena G, Su MY, Cha S, Filippi CG, Bota D, Baldi P, Poisson LM, Jain R, Chow D (2018) Deep-learning convolutional neural networks accurately classify genetic mutations in gliomas. AJNR Am J Neuroradiol 39:1201–1207
Eichinger P, Alberts E, Delbridge C, Trebeschi S, Valentinitsch A, Bette S, Huber T, Gempt J, Meyer B, Schlegel J, Zimmer C, Kirschke JS, Menze BH, Wiestler B (2017) Diffusion tensor image features predict IDH genotype in newly diagnosed WHO grade II/III gliomas. Sci Rep 7:13396
Han L, Kamdar MR (2018) MRI to MGMT: predicting methylation status in glioblastoma patients using convolutional recurrent neural networks. Pac Symp Biocomput 23:331–342
Han Y, Xie Z, Zang Y, Zhang S, Gu D, Zhou M, Gevaert O, Wei J, Li C, Chen H, Du J, Liu Z, Dong D, Tian J, Zhou D (2018) Non-invasive genotype prediction of chromosome 1p/19q co-deletion by development and validation of an MRI-based radiomics signature in lower-grade gliomas. J Neurooncol 140:297–306
Kong Z, Lin Y, Jiang C, Li L, Liu Z, Wang Y, Dai C, Liu D, Qin X, Wang Y, Liu Z, Cheng X, Tian J, Ma W (2019) (18)F-FDG-PET-based radiomics signature predicts MGMT promoter methylation status in primary diffuse glioma. Cancer Imaging 19:58
Korfiatis P, Kline TL, Coufalova L, Lachance DH, Parney IF, Carter RE, Buckner JC, Erickson BJ (2016) MRI texture features as biomarkers to predict MGMT methylation status in glioblastomas. Med Phys 43:2835–2844
Korfiatis P, Kline TL, Lachance DH, Parney IF, Buckner JC, Erickson BJ (2017) Residual deep convolutional neural network predicts MGMT methylation status. J Digit Imaging 30:622–628
Li Z, Wang Y, Yu J, Guo Y, Cao W (2017) Deep learning based radiomics (DLR) and its usage in noninvasive IDH1 prediction for low grade glioma. Sci Rep 7:5467
Li ZC, Bai H, Sun Q, Li Q, Liu L, Zou Y, Chen Y, Liang C, Zheng H (2018) Multiregional radiomics features from multiparametric MRI for prediction of MGMT methylation status in glioblastoma multiforme: a multicentre study. Eur Radiol 28:3640–3650
Liu X, Li Y, Li S, Fan X, Sun Z, Yang Z, Wang K, Zhang Z, Jiang T, Liu Y, Wang L, Wang Y (2019) IDH mutation-specific radiomic signature in lower-grade gliomas. Aging (Albany NY) 11:673–696
Lohmann P, Lerche C, Bauer EK, Steger J, Stoffels G, Blau T, Dunkl V, Kocher M, Viswanathan S, Filss CP, Stegmayr C, Ruge MI, Neumaier B, Shah NJ, Fink GR, Langen KJ, Galldiks N (2018) Predicting IDH genotype in gliomas using FET PET radiomics. Sci Rep 8:13328
Lu CF, Hsu FT, Hsieh KL, Kao YJ, Cheng SJ, Hsu JB, Tsai PH, Chen RJ, Huang CC, Yen Y, Chen CY (2018) Machine learning-based radiomics for molecular subtyping of gliomas. Clin Cancer Res 24:4429–4436
Shofty B, Artzi M, Ben Bashat D, Liberman G, Haim O, Kashanian A, Bokstein F, Blumenthal DT, Ram Z, Shahar T (2017) MRI radiomics analysis of molecular alterations in low-grade gliomas. Int J Comput Assist Radiol Surg. https://doi.org/10.1007/s11548-017-1691-5
Xi YB, Guo F, Xu ZL, Li C, Wei W, Tian P, Liu TT, Liu L, Chen G, Ye J, Cheng G, Cui LB, Zhang HJ, Qin W, Yin H (2017) Radiomics signature: a potential biomarker for the prediction of MGMT promoter methylation in glioblastoma. J Magn Reson Imaging. https://doi.org/10.1002/jmri.25860
Zhang X, Tian Q, Wang L, Liu Y, Li B, Liang Z, Gao P, Zheng K, Zhao B, Lu H (2018) Radiomics strategy for molecular subtype stratification of lower-grade glioma: detecting IDH and TP53 mutations based on multimodal MRI. J Magn Reson Imaging 48:916–926
Zhou H, Chang K, Bai HX, Xiao B, Su C, Bi WL, Zhang PJ, Senders JT, Vallieres M, Kavouridis VK, Boaro A, Arnaout O, Yang L, Huang RY (2019) Machine learning reveals multimodal MRI patterns predictive of isocitrate dehydrogenase and 1p/19q status in diffuse low- and high-grade gliomas. J Neurooncol 142:299–307
Zhou H, Vallieres M, Bai HX, Su C, Tang H, Oldridge D, Zhang Z, Xiao B, Liao W, Tao Y, Zhou J, Zhang P, Yang L (2017) MRI features predict survival and molecular markers in diffuse lower-grade gliomas. Neuro Oncol 19:862–870
Pyka T, Gempt J, Hiob D, Ringel F, Schlegel J, Bette S, Wester HJ, Meyer B, Forster S (2016) Textural analysis of pre-therapeutic [18F]-FET-PET and its correlation with tumor grade and patient survival in high-grade gliomas. Eur J Nucl Med Mol Imaging 43:133–141
Menze BH, Jakab A, Bauer S, Kalpathy-Cramer J, Farahani K, Kirby J, Burren Y, Porz N, Slotboom J, Wiest R, Lanczi L, Gerstner E, Weber MA, Arbel T, Avants BB, Ayache N, Buendia P, Collins DL, Cordier N, Corso JJ, Criminisi A, Das T, Delingette H, Demiralp C, Durst CR, Dojat M, Doyle S, Festa J, Forbes F, Geremia E, Glocker B, Golland P, Guo X, Hamamci A, Iftekharuddin KM, Jena R, John NM, Konukoglu E, Lashkari D, Mariz JA, Meier R, Pereira S, Precup D, Price SJ, Raviv TR, Reza SM, Ryan M, Sarikaya D, Schwartz L, Shin HC, Shotton J, Silva CA, Sousa N, Subbanna NK, Szekely G, Taylor TJ, Thomas OM, Tustison NJ, Unal G, Vasseur F, Wintermark M, Ye DH, Zhao L, Zhao B, Zikic D, Prastawa M, Reyes M, Van Leemput K (2015) The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans Med Imaging 34:1993–2024
Li ZC, Bai H, Sun Q, Zhao Y, Lv Y, Zhou J, Liang C, Chen Y, Liang D, Zheng H (2018) Multiregional radiomics profiling from multiparametric MRI: identifying an imaging predictor of IDH1 mutation status in glioblastoma. Cancer Med 7:5999–6009
Zhang B, Chang K, Ramkissoon S, Tanguturi S, Bi WL, Reardon DA, Ligon KL, Alexander BM, Wen PY, Huang RY (2017) Multimodal MRI features predict isocitrate dehydrogenase genotype in high-grade gliomas. Neuro Oncol 19:109–117
Kickingereder P, Isensee F, Tursunova I, Petersen J, Neuberger U, Bonekamp D, Brugnara G, Schell M, Kessler T, Foltyn M, Harting I, Sahm F, Prager M, Nowosielski M, Wick A, Nolden M, Radbruch A, Debus J, Schlemmer HP, Heiland S, Platten M, von Deimling A, van den Bent MJ, Gorlia T, Wick W, Bendszus M, Maier-Hein KH (2019) Automated quantitative tumour response assessment of MRI in neuro-oncology with artificial neural networks: a multicentre, retrospective study. Lancet Oncol 20:728–740
Shaver MM, Kohanteb PA, Chiou C, Bardis MD, Chantaduly C, Bota D, Filippi CG, Weinberg B, Grinband J, Chow DS, Chang PD (2019) Optimizing neuro-oncology imaging: a review of deep learning approaches for glioma imaging. Cancers (Basel) 11(6):E829. https://doi.org/10.3390/cancers11060829
Bakas S, Reyes M, Jakab A, Bauer S, Rempfler M, Crimi A, Shinohara RT, Berger C, Ha SM, Rozycki M (2018) Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the BRATS challenge (arXiv:1811.02629)
Isensee F, Kickingereder P, Wick W, Bendszus M, Maier-Hein KH (2018) Brain tumor segmentation and radiomics survival prediction: contribution to the BRATS 2017 challenge (arXiv:1802.10508)
Zhuge Y, Krauze AV, Ning H, Cheng JY, Arora BC, Camphausen K, Miller RW (2017) Brain tumor segmentation using holistically nested neural networks in MRI images. Med Phys 44:5234–5243
Wu YP, Lin YS, Wu WG, Yang C, Gu JQ, Bai Y, Wang MY (2017) Semiautomatic segmentation of glioma on mobile devices. J Healthc Eng 2017:8054939
Chamberlain MC (2011) Radiographic patterns of relapse in glioblastoma. J Neurooncol 101:319–323
Oppitz U, Maessen D, Zunterer H, Richter S, Flentje M (1999) 3D-recurrence-patterns of glioblastomas after CT-planned postoperative irradiation. Radiother Oncol 53:53–57
Syed M, Liermann J, Verma V, Bernhardt D, Bougatf N, Paul A, Rieken S, Debus J, Adeberg S (2018) Survival and recurrence patterns of multifocal glioblastoma after radiation therapy. Cancer Manag Res 10:4229–4235
Rapp M, Baernreuther J, Turowski B, Steiger HJ, Sabel M, Kamp MA (2017) Recurrence pattern analysis of primary glioblastoma. World Neurosurg 103:733–740
Minniti G, Amelio D, Amichetti M, Salvati M, Muni R, Bozzao A, Lanzetta G, Scarpino S, Arcella A, Enrici RM (2010) Patterns of failure and comparison of different target volume delineations in patients with glioblastoma treated with conformal radiotherapy plus concomitant and adjuvant temozolomide. Radiother Oncol 97:377–381
Cabrera AR, Kirkpatrick JP, Fiveash JB, Shih HA, Koay EJ, Lutz S, Petit J, Chao ST, Brown PD, Vogelbaum M, Reardon DA, Chakravarti A, Wen PY, Chang E (2016) Radiation therapy for glioblastoma: executive summary of an American society for radiation oncology evidence-based clinical practice guideline. Pract Radiat Oncol 6:217–225
Niyazi M, Brada M, Chalmers AJ, Combs SE, Erridge SC, Fiorentino A, Grosu AL, Lagerwaard FJ, Minniti G, Mirimanoff RO, Ricardi U, Short SC, Weber DC, Belka C (2016) ESTRO-ACROP guideline “target delineation of glioblastomas”. Radiother Oncol 118:35–42
Chan JL, Lee SW, Fraass BA, Normolle DP, Greenberg HS, Junck LR, Gebarski SS, Sandler HM (2002) Survival and failure patterns of high-grade gliomas after three-dimensional conformal radiotherapy. J Clin Oncol 20:1635–1642
Nakagawa K, Aoki Y, Fujimaki T, Tago M, Terahara A, Karasawa K, Sakata K, Sasaki Y, Matsutani M, Akanuma A (1998) High-dose conformal radiotherapy influenced the pattern of failure but did not improve survival in glioblastoma multiforme. Int J Radiat Oncol Biol Phys 40:1141–1149
Tanaka M, Ino Y, Nakagawa K, Tago M, Todo T (2005) High-dose conformal radiotherapy for supratentorial malignant glioma: a historical comparison. Lancet Oncol 6:953–960
Monjazeb AM, Ayala D, Jensen C, Case LD, Bourland JD, Ellis TL, McMullen KP, Chan MD, Tatter SB, Lesser GJ, Shaw EG (2012) A phase I dose escalation study of hypofractionated IMRT field-in-field boost for newly diagnosed glioblastoma multiforme. Int J Radiat Oncol Biol Phys 82:743–748
Piroth MD, Pinkawa M, Holy R, Klotz J, Schaar S, Stoffels G, Galldiks N, Coenen HH, Kaiser HJ, Langen KJ, Eble MJ (2012) Integrated boost IMRT with FET-PET-adapted local dose escalation in glioblastomas. Results of a prospective phase II study. Strahlenther Onkol 188:334–339
Piroth MD, Galldiks N, Pinkawa M, Holy R, Stoffels G, Ermert J, Mottaghy FM, Shah NJ, Langen KJ, Eble MJ (2016) Relapse patterns after radiochemotherapy of glioblastoma with FET PET-guided boost irradiation and simulation to optimize radiation target volume. Radiat Oncol 11:87
Weber DC, Casanova N, Zilli T, Buchegger F, Rouzaud M, Nouet P, Vees H, Ratib O, Dipasquale G, Miralbell R (2009) Recurrence pattern after [(18)F]fluoroethyltyrosine-positron emission tomography-guided radiotherapy for high-grade glioma: a prospective study. Radiother Oncol 93:586–592
Bette S, Huber T, Gempt J, Boeckh-Behrens T, Wiestler B, Kehl V, Ringel F, Meyer B, Zimmer C, Kirschke JS (2017) Local fractional anisotropy is reduced in areas with tumor recurrence in glioblastoma. Radiology 283:499–507
Elson A, Paulson E, Bovi J, Siker M, Schultz C, Laviolette PS (2015) Evaluation of pre-radiotherapy apparent diffusion coefficient (ADC): patterns of recurrence and survival outcomes analysis in patients treated for glioblastoma multiforme. J Neurooncol 123:179–188
Esmaeili M, Stensjoen AL, Berntsen EM, Solheim O, Reinertsen I (2018) The direction of tumour growth in glioblastoma patients. Sci Rep 8:1199
Jayamanne D, Wheeler H, Brazier D, Newey A, Kastelan M, Guo L, Back M (2018) Predicting patterns of failure in temporal lobe GBMs: possible implications on radiotherapy treatment portals. Radiat Oncol 13:133
Akbari H, Macyszyn L, Da X, Bilello M, Wolf RL, Martinez-Lage M, Biros G, Alonso-Basanta M, O’Rourke DM, Davatzikos C (2016) Imaging surrogates of infiltration obtained via multiparametric imaging pattern analysis predict subsequent location of recurrence of glioblastoma. Neurosurgery 78:572–580
Akbari H, Macyszyn L, Da X, Wolf RL, Bilello M, Verma R, O’Rourke DM, Davatzikos C (2014) Pattern analysis of dynamic susceptibility contrast-enhanced MR imaging demonstrates peritumoral tissue heterogeneity. Radiology 273:502–510
Rathore S, Akbari H, Doshi J, Shukla G, Rozycki M, Bilello M, Lustig R, Davatzikos C (2018) Radiomic signature of infiltration in peritumoral edema predicts subsequent recurrence in glioblastoma: implications for personalized radiotherapy planning. J Med Imaging (Bellingham) 5:21219
Kickingereder P, Burth S, Wick A, Gotz M, Eidel O, Schlemmer HP, Maier-Hein KH, Wick W, Bendszus M, Radbruch A, Bonekamp D (2016) Radiomic profiling of glioblastoma: identifying an imaging predictor of patient survival with improved performance over established clinical and radiologic risk models. Radiology 280:880–889
Prasanna P, Patel J, Partovi S, Madabhushi A, Tiwari P (2017) Radiomic features from the peritumoral brain parenchyma on treatment-naive multi-parametric MR imaging predict long versus short-term survival in glioblastoma multiforme: preliminary findings. Eur Radiol 27:4188–4197
Jain R, Poisson LM, Gutman D, Scarpace L, Hwang SN, Holder CA, Wintermark M, Rao A, Colen RR, Kirby J, Freymann J, Jaffe CC, Mikkelsen T, Flanders A (2014) Outcome prediction in patients with glioblastoma by using imaging, clinical, and genomic biomarkers: focus on the nonenhancing component of the tumor. Radiology 272:484–493
Kickingereder P, Neuberger U, Bonekamp D, Piechotta PL, Gotz M, Wick A, Sill M, Kratz A, Shinohara RT, Jones DTW, Radbruch A, Muschelli J, Unterberg A, Debus J, Schlemmer HP, Herold-Mende C, Pfister S, von Deimling A, Wick W, Capper D, Maier-Hein KH, Bendszus M (2018) Radiomic subtyping improves disease stratification beyond key molecular, clinical, and standard imaging characteristics in patients with glioblastoma. Neuro Oncol 20:848–857
Kim JY, Yoon MJ, Park JE, Choi EJ, Lee J, Kim HS (2019) Radiomics in peritumoral non-enhancing regions: fractional anisotropy and cerebral blood volume improve prediction of local progression and overall survival in patients with glioblastoma. Neuroradiology. https://doi.org/10.1007/s00234-019-02255-4
Mohsen LA, Shi V, Jena R, Gillard JH, Price SJ (2013) Diffusion tensor invasive phenotypes can predict progression-free survival in glioblastomas. Br J Neurosurg 27:436–441
Galldiks N, Kocher M, Langen KJ (2017) Pseudoprogression after glioma therapy: an update. Expert Rev Neurother 17:1109–1115
Young RJ, Gupta A, Shah AD, Graber JJ, Zhang Z, Shi W, Holodny AI, Omuro AM (2011) Potential utility of conventional MRI signs in diagnosing pseudoprogression in glioblastoma. Neurology 76:1918–1924
Wen PY, Macdonald DR, Reardon DA, Cloughesy TF, Sorensen AG, Galanis E, Degroot J, Wick W, Gilbert MR, Lassman AB, Tsien C, Mikkelsen T, Wong ET, Chamberlain MC, Stupp R, Lamborn KR, Vogelbaum MA, van den Bent MJ, Chang SM (2010) Updated response assessment criteria for high-grade gliomas: response assessment in neuro-oncology working group. J Clin Oncol 28:1963–1972
Galldiks N, Dunkl V, Stoffels G, Hutterer M, Rapp M, Sabel M, Reifenberger G, Kebir S, Dorn F, Blau T, Herrlinger U, Hau P, Ruge MI, Kocher M, Goldbrunner R, Fink GR, Drzezga A, Schmidt M, Langen KJ (2015) Diagnosis of pseudoprogression in patients with glioblastoma using O‑(2-[18F]fluoroethyl)-L-tyrosine PET. Eur J Nucl Med Mol Imaging 42:685–695
Patel P, Baradaran H, Delgado D, Askin G, Christos P, John Tsiouris A, Gupta A (2017) MR perfusion-weighted imaging in the evaluation of high-grade gliomas after treatment: a systematic review and meta-analysis. Neuro Oncol 19:118–127
Filss CP, Cicone F, Shah NJ, Galldiks N, Langen KJ (2017) Amino acid PET and MR perfusion imaging in brain tumours. Clin Transl Imaging 5:209–223
Hu X, Wong KK, Young GS, Guo L, Wong ST (2011) Support vector machine multiparametric MRI identification of pseudoprogression from tumor recurrence in patients with resected glioblastoma. J Magn Reson Imaging 33:296–305
Kim JY, Park JE, Jo Y, Shim WH, Nam SJ, Kim JH, Yoo RE, Choi SH, Kim HS (2019) Incorporating diffusion- and perfusion-weighted MRI into a radiomics model improves diagnostic performance for pseudoprogression in glioblastoma patients. Neuro Oncol 21:404–414
Kebir S, Khurshid Z, Gaertner FC, Essler M, Hattingen E, Fimmers R, Scheffler B, Herrlinger U, Bundschuh RA, Glas M (2017) Unsupervised consensus cluster analysis of [18F]-fluoroethyl-L-tyrosine positron emission tomography identified textural features for the diagnosis of pseudoprogression in high-grade glioma. Oncotarget 8:8294–8304
Lohmann P, Elahmadawy MA, Werner J, Rapp M, Ceccon G, Fink GR, Shah NJ, Langen K, Galldiks N (2019) OS9.6 diagnosis of pseudoprogression using FET PET radiomics. Neuro Oncol 21:iii19–iii19
Jang BS, Jeon SH, Kim IH, Kim IA (2018) Prediction of pseudoprogression versus progression using machine learning algorithm in glioblastoma. Sci Rep 8:12516
Bacchi S, Zerner T, Dongas J, Asahina AT, Abou-Hamden A, Otto S, Oakden-Rayner L, Patel S (2019) Deep learning in the detection of high-grade glioma recurrence using multiple MRI sequences: a pilot study. J Clin Neurosci 70:11–13
Akbari H, Rathore S, Bakas S, Nasrallah MP, Shukla G, Mamourian E, Rozycki M, Bagley SJ, Rudie JD, Flanders AE, Dicker AP, Desai AS, O’Rourke DM, Brem S, Lustig R, Mohan S, Wolf RL, Bilello M, Martinez-Lage M, Davatzikos C (2020) Histopathology-validated machine learning radiographic biomarker for noninvasive discrimination between true progression and pseudo-progression in glioblastoma. Cancer. https://doi.org/10.1002/cncr.32790
Li M, Tang H, Chan MD, Zhou X, Qian X (2020) DC-AL GAN: pseudoprogression and true tumor progression of glioblastoma multiform image classification based on DCGAN and Alexnet. Med Phys 47:1139–1150
Ohgaki H, Kleihues P (2005) Epidemiology and etiology of gliomas. Acta Neuropathol 109:93–108
Platta CS, Khuntia D, Mehta MP, Suh JH (2010) Current treatment strategies for brain metastasis and complications from therapeutic techniques: a review of current literature. Am J Clin Oncol 33:398–407
Qian Z, Li Y, Wang Y, Li L, Li R, Wang K, Li S, Tang K, Zhang C, Fan X, Chen B, Li W (2019) Differentiation of glioblastoma from solitary brain metastases using radiomic machine-learning classifiers. Cancer Lett 451:128–135
Artzi M, Bressler I, Ben Bashat D (2019) Differentiation between glioblastoma, brain metastasis and subtypes using radiomics analysis. J Magn Reson Imaging 50:519–528
National Comprehensive Cancer Network (2019) Central nervous system cancers—version 3.2019. NCCN clinical practice guidelines in oncology (NCCN guidelines) (Version 3.2019)
Liu Y, Stojadinovic S, Hrycushko B, Wardak Z, Lau S, Lu W, Yan Y, Jiang SB, Zhen X, Timmerman R, Nedzi L, Gu X (2017) A deep convolutional neural network-based automatic delineation strategy for multiple brain metastases stereotactic radiosurgery. PLoS ONE 12:e185844
Charron O, Lallement A, Jarnet D, Noblet V, Clavier JB, Meyer P (2018) Automatic detection and segmentation of brain metastases on multimodal MR images with a deep convolutional neural network. Comput Biol Med 95:43–54
Brown JM, Carlson DJ, Brenner DJ (2014) The tumor radiobiology of SRS and SBRT: are more than the 5 Rs involved? Int J Radiat Oncol Biol Phys 88:254–262
Kocher M, Voges J, Treuer H, Sturm V, Müller R‑P (2000) Reduced response rate of necrotic brain metastases to radiosurgery. In: Kondziolka D (ed) Radiosurgery 1999. Karger, Basel, pp 240–246
Della Seta M, Collettini F, Chapiro J, Angelidis A, Engeling F, Hamm B, Kaul D (2019) A 3D quantitative imaging biomarker in pre-treatment MRI predicts overall survival after stereotactic radiation therapy of patients with a singular brain metastasis. Acta Radiol 60:1496–1503
Tini P, Nardone V, Pastina P, Battaglia G, Vinciguerra C, Carfagno T, Rubino G, Carbone SF, Sebaste L, Cerase A, Federico A, Pirtoli L (2017) Perilesional edema in brain metastasis from non-small cell lung cancer (NSCLC) as predictor of response to radiosurgery (SRS). Neurol Sci 38:975–982
Cha YJ, Jang WI, Kim MS, Yoo HJ, Paik EK, Jeong HK, Youn SM (2018) Prediction of response to stereotactic radiosurgery for brain metastases using convolutional neural networks. Anticancer Res 38:5437–5445
Mouraviev A, Detsky J, Sahgal A, Ruschin M, Lee YK, Karam I, Heyn C, Stanisz GJ, Martel AL (2020) Use of radiomics for the prediction of local control of brain metastases after stereotactic radiosurgery. Neuro Oncol. https://doi.org/10.1093/neuonc/noaa007
Sneed PK, Mendez J, Vemer-van den Hoek JG, Seymour ZA, Ma L, Molinaro AM, Fogh SE, Nakamura JL, McDermott MW (2015) Adverse radiation effect after stereotactic radiosurgery for brain metastases: incidence, time course, and risk factors. J Neurosurg 123:373–386
Kondziolka D, Shin SM, Brunswick A, Kim I, Silverman JS (2015) The biology of radiosurgery and its clinical applications for brain tumors. Neuro Oncol 17:29–44
Peng L, Parekh V, Huang P, Lin DD, Sheikh K, Baker B, Kirschbaum T, Silvestri F, Son J, Robinson A, Huang E, Ames H, Grimm J, Chen L, Shen C, Soike M, McTyre E, Redmond K, Lim M, Lee J, Jacobs MA, Kleinberg L (2018) Distinguishing true progression from radionecrosis after stereotactic radiation therapy for brain metastases with machine learning and radiomics. Int J Radiat Oncol Biol Phys 102:1236–1243
Spitzer M, Lorkowski S, Cullen P, Sczyrba A, Fuellen G (2006) IsoSVM—distinguishing isoforms and paralogs on the protein level. BMC Bioinformatics 7:110
Zhang Z, Yang J, Ho A, Jiang W, Logan J, Wang X, Brown PD, McGovern SL, Guha-Thakurta N, Ferguson SD, Fave X, Zhang L, Mackin D, Court LE, Li J (2018) A predictive model for distinguishing radiation necrosis from tumour progression after gamma knife radiosurgery based on radiomic features from MR images. Eur Radiol 28:2255–2263
Ceccon G, Lohmann P, Stoffels G, Judov N, Filss CP, Rapp M, Bauer E, Hamisch C, Ruge MI, Kocher M, Kuchelmeister K, Sellhaus B, Sabel M, Fink GR, Shah NJ, Langen KJ, Galldiks N (2017) Dynamic O‑(2-18F-fluoroethyl)-L-tyrosine positron emission tomography differentiates brain metastasis recurrence from radiation injury after radiotherapy. Neuro Oncol 19:281–288
Lohmann P, Stoffels G, Ceccon G, Rapp M, Sabel M, Filss CP, Kamp MA, Stegmayr C, Neumaier B, Shah NJ, Langen KJ, Galldiks N (2017) Radiation injury vs. recurrent brain metastasis: combining textural feature radiomics analysis and standard parameters may increase (18)F-FET PET accuracy without dynamic scans. Eur Radiol 27:2916–2927
Lohmann P, Kocher M, Ceccon G, Bauer EK, Stoffels G, Viswanathan S, Ruge MI, Neumaier B, Shah NJ, Fink GR, Langen KJ, Galldiks N (2018) Combined FET PET/MRI radiomics differentiates radiation injury from recurrent brain metastasis. Neuroimage Clin 20:537–542
Zwanenburg A, Vallieres M, Abdalah MA, Aerts H, Andrearczyk V, Apte A, Ashrafinia S, Bakas S, Beukinga RJ, Boellaard R, Bogowicz M, Boldrini L, Buvat I, Cook GJR, Davatzikos C, Depeursinge A, Desseroit MC, Dinapoli N, Dinh CV, Echegaray S, El Naqa I, Fedorov AY, Gatta R, Gillies RJ, Goh V, Gotz M, Guckenberger M, Ha SM, Hatt M, Isensee F, Lambin P, Leger S, Leijenaar RTH, Lenkowicz J, Lippert F, Losnegard A, Maier-Hein KH, Morin O, Muller H, Napel S, Nioche C, Orlhac F, Pati S, Pfaehler EAG, Rahmim A, Rao AUK, Scherer J, Siddique MM, Sijtsema NM, Socarras Fernandez J, Spezi E, Steenbakkers R, Tanadini-Lang S, Thorwarth D, Troost EGC, Upadhaya T, Valentini V, van Dijk LV, van Griethuysen J, van Velden FHP, Whybra P, Richter C, Lock S (2020) The image biomarker standardization initiative: standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology. https://doi.org/10.1148/radiol.2020191145:191145
Funding
Supported by the Deutsche Forschungsgemeinschaft (DFG), project number 428090865 (N.G. and P.L.)
Funding
Open Access funding provided by Projekt DEAL.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
M. Kocher, M.I. Ruge, N. Galldiks, and P. Lohmann declare that they have no competing interests.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kocher, M., Ruge, M.I., Galldiks, N. et al. Applications of radiomics and machine learning for radiotherapy of malignant brain tumors. Strahlenther Onkol 196, 856–867 (2020). https://doi.org/10.1007/s00066-020-01626-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00066-020-01626-8