Abstract
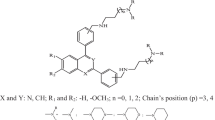
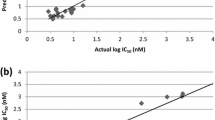
The spread of antimalarial drug resistance and lack of effective vaccines into consideration drive significant role to develop novel antimalarial agents for the treatment of malaria. The Plasmodium falciparum deoxyuridine 5′-triphosphate nucleotidohydrolase (dUTPase) has been identified as a promising target for new drug development. In this study, we report a receptor-dependent mixed 2D- and 3D-QSAR model using the LQTA-QSAR methodology on a series of dUTPase inhibitors. QSAR models were constructed by a training set of 55 compounds with specific stereochemistry and an external set of 18. The model satisfied a set of rigorous validation criteria and performed well in the prediction of an external test set. The proposed model also checked for free from chance correlation, reliability, and robustness by y-randomization and leave-N-out tests. Final model provided the following statistics (R 2 = 0.85, Q 2LOO = 0.78 and Q 2ext = 0.76) with only 12 descriptors and 3 latent variables. We showed that a combination of 2D classic and 3D molecular field descriptors could lead to relevant QSAR models, that in comparison with model solely generated by 3D descriptors has improved and illustrated more robustness and prediction ability. Visualization of the descriptors of the best model helps us to interpret the model from the chemical point of view, supporting the applicability of this new approach in rational drug design.



Similar content being viewed by others
References
Accelrys (2002) ViewerLite. San Diego, CA, USA
Andrade CH, Pasqualoto KFM, Ferreira EI, Hopfinger AJ (2010) 4D-QSAR: perspectives in drug design. Molecules 15:3281–3294
Baragaña B, McCarthy O, Sánchez P et al (2011) β-Branched acyclic nucleoside analogues as inhibitors of Plasmodium falciparum dUTPase. Bioorgan Med Chem 19:2378–2391
Barbosa EG, Ferreira MMC (2012) Digital filters for molecular interaction field descriptors. Mol Inf 31:75–84
Berman HM, Westbrook J, Feng Z et al (2000) The Protein Data Bank. Nucleic Acids Res 28:235–242
Delphine CB, David MR, Jan HJ (2008) Very fast prediction and rationalization of pKa values for protein–ligand complexes. Proteins 73:765–783
Eriksson L, Jaworska J, Andrew P et al (2003) Methods for reliability and uncertainty assessment and for applicability evaluations of classification- and regression-based QSARs. Environ Health Perspect 111:1361–1375
Fourches D, Muratov E, Tropsha A (2010) Trust, but verify: on the importance of chemical structure curation in cheminformatics and QSAR modeling research. J Chem Inf Model 50:1189–1204
Gadsden MH, McIntosh EM, Game JC et al (1993) dUTP pyrophosphatase is an essential enzyme in Saccharomyces cerevisiae. EMBO J 12:4425–4431
Gramatica P (2007) Principles of QSAR models validation: internal and external. QSAR Comb Sci 26:694–701
Halgren T (1996) Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. J Comput Chem 17(5–6):490–519
Hampton SE, Baragaña B, Schipani A et al (2011) Design, synthesis, and evaluation of 5′-diphenyl nucleoside analogues as inhibitors of the Plasmodium falciparum dUTPase. ChemMedChem 6:1816–1831
Hanwell M, Curtis D, Lonie D et al (2012) Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. J Chem Inf 4:17
Kiralj R, Ferreira MMC (2009) Basic validation procedures for regression models in QSAR and QSPR studies: theory and application. J Brazil Chem Soc 20:770–787
Kiralj R, Ferreira MMC (2010) Is your QSAR/QSPR descriptor real or trash? J Chemom 24:681–693
Kubinyi H (1997) QSAR and 3D QSAR in drug design Part 1: methodology. Drug Discov Today 2:457–467
Martins JPA, Barbosa EG, Pasqualoto KFM, Ferreira MMC (2009) LQTA-QSAR: a new 4D-QSAR methodology. J Chem Inf Model 49:1428–1436
McCarthy O, Musso-Buendia A, Kaiser M et al (2009) Design, synthesis and evaluation of novel uracil acetamide derivatives as potential inhibitors of Plasmodium falciparum dUTP nucleotidohydrolase. Eur J Med Chem 44:678–688
Nguyen C, Kasinathan G, Leal-Cortijo I et al (2005) Deoxyuridine triphosphate nucleotidohydrolase as a potential antiparasitic drug target. J Med Chem 48:5942–5954
Nguyen C, Ruda GF, Schipani A et al (2006) Acyclic nucleoside analogues as inhibitors of Plasmodium falciparum dUTPase. J Med Chem 49:4183–4195
O’Boyle N, Banck M, James C, Morley C et al (2011) Open Babel: an open chemical toolbox. J Chem Inf 3:33
Olliaro PL, Yuthavong Y (1999) An overview of chemotherapeutic targets for antimalarial drug discovery. Pharmacol Ther 81:91–110
Pettersen EF, Goddard TD, Huang CC, Couch GS et al (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612
Richie TL, Saul A (2002) Progress and challenges for malaria vaccines. Nature 415:694–701
Ruda GF, Nguyen C, Ziemkowski P et al (2011) Modified 5′-trityl nucleosides as inhibitors of Plasmodium falciparum dUTPase. ChemMedChem 6(2):309–320
Teófilo RF, Martins JPA et al (2009) Sorting variables by using informative vectors as a strategy for feature selection in multivariate regression. J Chemom 23:32–48
Tetko I, Gasteiger J, Todeschini R, Mauri A et al (2005) Virtual computational chemistry laboratory: design and description. J Comput Aided Mol Des 19:453–463
Tosco P, Balle T (2011) Open3DQSAR: a new open-source software aimed at high-throughput chemometric analysis of molecular interaction fields. J Mol Model 17:201–208
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31:455–461
Tseng YJ, Hopfinger AJ, Esposito EX (2011) The great descriptor melting pot: mixing descriptors for the common good of QSAR models. J Comput Aided Mol Des 26:39–43
Whittingham JL, Leal I, Nguyen C, Kasinathan G et al (2005) dUTPase as a platform for antimalarial drug design: structural basis for the selectivity of a class of nucleoside inhibitors. Structure 13(2):329–338
World Health Organization (2011) Global report on antimalarial drug efficacy and drug resistance: 2000–2010. W. L. C.-i.-P. Data. Geneva
World Health Organization (2011) World Malaria Report 2011. W. L. C.-i.-P. Data. Geneva
Yufeng JT, Anton JH, Emilio XE (2011) The great descriptor melting pot: mixing descriptors for the common good of QSAR models. J Comput Aided Mol Des 26:39–43
Acknowledgments
The authors wish to thank the department of pharmacy of the Federal University of Rio Grande do Norte, in the name of Prof. Nilsen Carvalho Fernandes de Oliveira Filho, for providing the required resources so that this work could be developed. I also wish to thank PPgCF/UFRN for the English revision.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
de Araújo Santos, R.A., Braz, C.A., Ghasemi, J.B. et al. Mixed 2D–3D-LQTA-QSAR study of a series of Plasmodium falciparum dUTPase inhibitors. Med Chem Res 24, 1098–1111 (2015). https://doi.org/10.1007/s00044-014-1189-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00044-014-1189-4