Abstract
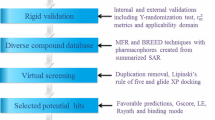
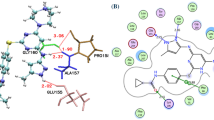
The ataxia telangiectasia mutated and Rad3 related (ATR) protein kinase is one of the apical regulators in the DNA damaging response signaling pathways. Inhibition of ATR kinase may greatly potentiates the cyto-toxicity by DNA damaging agents to the tumor cells while, have minimum effect on normal cells. In this article, the impact of ligand conformation on the three-dimensional quantitative structure–activity relationship (3D-QSAR) model of a series of 3-amino-6-arylpyrazines as ATR kinase inhibitors was investigated. We employed molecular dynamics (MDs) simulations to get the dynamic active conformations (DACs) of the compounds in the ATP-binding site of ATR kinase. As a result, the model based on the DACs extracted from the first nanosecond MD simulation is superior to that using static active conformations from docking with r 2 = 0.917; \(q_{\text{LOO}}^{2}\) = 0.880; standard error of estimate [SEE] = 0.259; F = 347.29; \(r_{\text{pred}}^{2}\) = 0.923; SEEpred = 0.301. Our results highlight the importance of incorporating DACs of ligand using MD simulation in 3D-QSAR studies. This study may also provide useful information to rationalize the design of novel ATR kinase inhibitors.








Similar content being viewed by others
References
AbdulHameed MD, Hamza A, Liu JJ, Zhan CG (2008) Combined 3D-QSAR modeling and molecular docking study on indolinone derivatives as inhibitors of 3-phosphoinositide-dependent protein kinase-1. J Chem Inf Model 48:1760–1772
Aggarwal S, Thareja S, Bhardwaj TR, Kumar M (2010a) Self-organizing molecular field analysis on pregnane derivatives as human steroidal 5alpha-reductase inhibitors. Steroids 75:411–418
Aggarwal S, Thareja S, Bhardwaj T, Kumar M (2010b) 3D-QSAR studies on unsaturated 4-azasteroids as human 5α-reductase inhibitors: a self organizing molecular field analysis approach. Eur J Med Chem 45:476–481
Bolt J, Vo QN, Kim WJ, McWhorter AJ, Thomson J, Hagensee ME, Friedlander P, Brown KD, Gilbert J (2005) The ATM/p53 pathway is commonly targeted for inactivation in squamous cell carcinoma of the head and neck (SCCHN) by multiple molecular mechanisms. Oral Oncol 41:1013–1020
Borst P, Rottenberg S, Jonkers J (2008) How do real tumors become resistant to cisplatin? Cell Cycle 7:1353–1359
Bouaziz-Terrachet S, Terrachet R, Taïri-Kellou S (2013) Receptor and ligand-based 3D-QSAR study on a series of nonsteroidal anti-inflammatory drugs. Med Chem Res 22:1529–1537
Charrier J-D, Durrant SJ, Golec JMC, Kay DP, Knegtel RMA, MacCormick S, Mortimore M, O’Donnell ME, Pinder JL, Reaper PM, Rutherford AP, Wang PSH, Young SC, Pollard JR (2011) Discovery of potent and selective inhibitors of ataxia telangiectasia mutated and Rad3 related (ATR) protein kinase as potential anticancer agents. J Med Chem 54:2320–2330
Cimprich KA, Cortez D (2008) ATR: an essential regulator of genome integrity. Nat Rev Cell Biol 9:616–627
Cramer RD, Patterson DE, Bunce JD (1988) Comparative molecular field analysis (CoMFA). 1. Effect of shape on binding of steroids to carrier proteins. J Am Chem Soc 110:5959–5967
Ding L, Getz G, Wheeler DA et al (2008) Somatic mutations affect key pathways in lung adenocarcinoma. Nature 455:1069–1075
Discovery Studio, version 2.5, Accelrys Inc., San Diego, CA (2011)
Einhorn LH (2002) Curing metastatic testicular cancer. Proc Natl Acad Sci USA 99:4592–4595
Fiser A, Do RK, Sali A (2000) Modeling of loops in protein structures. Protein Sci 9:1753–1773
Frish MJ, Trucks GW, Schlegel HB et al (2004) GAUSSIAN 03, Revision D. 02. Gaussian, Inc., Wallingford
Galeazzi R (2009) Molecular dynamics as a tool in rational drug design: current status and some major applications. Curr Comput Aided Drug Des 5:225–240
Greenman C, Stephens P, Smith R et al (2007) Patterns of somatic mutation in human cancer genomes. Nature 446:153–158
Hou TJ, Wang JM, Li YY, Wang W (2011) Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J Chem Inf Model 51:69–82
Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graph 14:33–38
Iribarne F, Paulino M, Aguilera S, Tapia O (2009) Assaying phenothiazine derivatives as trypanothione reductase and glutathione reductase inhibitors by theoretical docking and molecular dynamics studies. J Mol Graph Model 28:371–381
Jiang H, Reinhardt HC, Bartkova J, Tommiska J, Blomqvist C, Nevanlinna H, Bartek J, Yaffe MB, Hemann MT (2009) The combined status of ATM and p53 link tumor development with therapeutic response. Genes Dev 23:1895–1909
Kastan MB, Bartek J (2004) Cell-cycle checkpoints and cancer. Nature 432:316–323
Kumar R, Kumar M (2013) 3D-QSAR CoMFA and CoMSIA studies for design of potent human steroid 5α-reductase inhibitors. Med Chem Res 22:105–114
Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26:283–291
Li MY, Du LP, Wu B, Xia L (2003) Self-organizing molecular field analysis on alpha (1a)-adrenoceptor dihydropyridine antagonists. Bioorg Med Chem 11:3945–3951
Li SL, He MY, Du HG (2011a) 3D-QSAR studies on a series of dihydroorotate dehydrogenase inhibitors: analogues of the active metabolite of leflunomide. Int J Mol Sci 12:2982–2993
Li Z, Zhou M, Wu F, Li R, Ding ZY (2011b) Self-organizing molecular field analysis on human β-secretase nonpeptide inhibitors: 5,5-disubstituted aminohydantoins. Eur J Med Chem 46:58–64
Lopez-Contreras AJ, Fernandez-Capetillo O (2010) The ATR barrier to replication-born DNA damage. DNA Repair 9:1249–1255
Nishida H, Tatewaki N, Nakajima Y, Magara T, Ko KM, Hamamori Y, Konishi T (2009) Inhibition of ATR protein kinase activity by schisandrin B in DNA damage response. Nucleic Acids Res 37:5678–5689
Ojha PK, Mitra I, Das RN, Roy K (2011) Further exploring r 2m metrics for validation of QSPR models. Chemom Intell Lab 107:194–205
Olaussen KA, Dunant A, Fouret P, Brambilla E, André F, Haddad V, Taranchon E, Filipits M, Pirker R, Popper HH, Stahel R, Sabatier L, Pignon J-P, Tursz T, Le Chevalier T, Soria J-C (2006) DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N Engl J Med 355:983–991
Oliver TG, Mercer KL, Sayles LC, Burke JR, Mendus D, Lovejoy KS, Cheng M-H, Subramanian A, Mu D, Powers S, Crowley D, Bronson RT, Whittaker CA, Bhutkar A, Lippard SJ et al (2010) Chronic cisplatin treatment promotes enhanced damage repair and tumor progression in a mouse model of lung cancer. Genes Dev 24:837–852
Peasland A, Wang LZ, Rowling E, Kyle S, Chen T, Hopkins A, Cliby WA, Sarkaria J, Beale G, Edmondson RJ, Curtin NJ (2011) Identification and evaluation of a potent novel ATR inhibitor, NU6027, in breast and ovarian cancer cell lines. Br J Cancer 105:372–381
Reaper PM, Griffiths MR, Long JM, Charrier JD, MacCormick S, Charlton PA, Golec JMC, Pollard JR (2011) Selective killing of ATM- or p53-deficient cancer cells through inhibition of ATR. Nat Chem Biol 7:428–430
Robinson DD, Winn PJ, Lyne PD, Richards WG (1999) Self-organizing molecular field analysis: a tool for structure–activity studies. J Med Chem 42:573–583
Roy PP, Paul S, Mitra I, Roy K (2009) On two novel parameters for validation of predictive QSAR models. Molecules 14:1660–1701
Roy K, Mitra I, Kar S, Ojha PK, Das RN, Kabir H (2012) Comparative studies on some metrics for external validation of QSPR models. J Chem Inf Model 52:396–408
Sangster-Guity N, Conrad BH, Papadopoulos N, Bunz F (2011) ATR mediates cisplatin resistance in a p53 genotype-specific manner. Oncogene 30:2526–2533
Sarkaria JN, Busby EC, Tibbetts RS, Roos P, Taya Y, Karnitz LM, Abraham RT (1999) Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res 59:4375–4382
Shen MY, Sali A (2006) Statistical potential for assessment and prediction of protein structures. Protein Sci 15:2507–2524
Sibanda BL, Chirgadze DY, Blundell TL (2010) Crystal structure of DNA-PKcs reveals a large open-ring cradle comprised of HEAT repeats. Nature 463:118–121
Sinha V, Thareja S, Kumar M (2012) Self-organizing molecular field analysis of NSAIDs: assessment of pharmacokinetic and physicochemical properties using 3D-QSPkR approach. Eur J Med Chem 53:76–82
Stewart Computational Chemistry (2008) Colorado Springs. http://OpenMOPAC.net. Accessed 1 July 2010
Thareja S, Aggarwal S, Bhardwaj TR, Kumar M (2009) Self organizing molecular field analysis on a series of human 5 alpha-reductase inhibitors: unsaturated 3-carboxysteroid. Eur J Med Chem 44:4920–4925
Thareja S, Aggarwal S, Bhardwaj T, Kumar M (2010) Self-organizing molecular field analysis of 2,4-thiazolidinediones: a 3D-QSAR model for the development of human PTP1B inhibitors. Eur J Med Chem 45:2537–2546
The Cambridge Crystallographic Data Centre, Cambridge (2010)
Toledo LI, Murga M, Zur R, Soria R, Rodriguez A, Martinez S, Oyarzabal J, Pastor J, Bischoff JR, Fernandez-Capetillo O (2011) A cell-based screen identifies ATR inhibitors with synthetic lethal properties for cancer-associated mutations. Nat Struct Mol Biol 18:721–727
Viktor H, Robert A, Asim O, Bentley S, Adrian R, Carlos S (2006) Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins 65:712–725
Walker EH, Pacold ME, Perisic O, Stephens L, Hawkins PT, Wymann MP, Williams RL (2000) Structural determinants of phosphoinositide 3-kinase inhibition by wortmannin, LY294002, quercetin, myricetin, and staurosporine. Mol Cell 6:909–919
Wang J, Romain MW, James WC, Peter AK, David AC (2004) Development and testing of a general Amber force field. J Comput Chem 25:1157–1174
Wang J, Wang W, Kollman P, Case D (2005) Antechamber, an accessory software package for molecular mechanical calculations. J Comput Chem 25:1157–1174
Acknowledgments
The authors are grateful for the support of the National Science Foundation of China (30901743), the National Key Program of China (2012ZX09103101-022), the State Key Laboratory of Drug Research, and Doctoral Fund of Ministry of Education.
Author information
Authors and Affiliations
Corresponding author
Additional information
Hao Luo and Jianyou Shi have contributed equally to this study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Luo, H., Shi, J., Lu, L. et al. Molecular dynamics-based self-organizing molecular field analysis on 3-amino-6-arylpyrazines as the ataxia telangiectasia mutated and Rad3 related (ATR) protein kinase inhibitors. Med Chem Res 23, 747–758 (2014). https://doi.org/10.1007/s00044-013-0665-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00044-013-0665-6