Abstract
Multiple myeloma (MM) is the second most common hematological tumor in adults. Immunomodulatory drugs (IMiDs), such as thalidomide and lenalidomide (Len), are effective drugs for the treatment of multiple myeloma. Len can recruit IKZF1 and IKZF3 to cereblon (CRBN), a substrate receptor of the cullin 4-RING E3 ligase (CRL4), promote their ubiquitination and degradation, and finally inhibit the proliferation of myeloma cells. However, MM patients develop resistance to IMiDs over time, leading to disease recurrence and deterioration. To explore the possible approaches that may enhance the sensitivity of IMiDs to MM, in this study, we used the proximity labeling technique TurboID and quantitative proteomics to identify Lys-63-specific deubiquitinase BRCC36 as a CRBN-interacting protein. Biochemical experiments demonstrated that BRCC36 in the BRISC complex protects CRBN from lysosomal degradation by specifically cleaving the K63-linked polyubiquitin chain on CRBN. Further studies found that a small-molecule compound SHIN1, which binds to BRISC complex subunit SHMT2, can upregulate CRBN by elevating BRCC36. The combination of SHIN1 and Len can further increase the sensitivity of MM cells to IMiDs. Therefore, this study provides the basis for the exploration of a possible strategy for the SHIN1 and Len combination treatment for MM.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Multiple myeloma (MM) is the second most frequently occurring hematologic malignancy [1]. Proteasome inhibitors such as bortezomib, immunomodulatory drugs (IMiDs) such as lenalidomide (Len), pomalidomide (Pom), anti-CD38 antibodies, and corticosteroids have been approved by the FDA for the treatment of myeloma [2,3,4]. Although these emerging new drugs have dramatically improved the overall survival of MM patients, the frequent occurrence of drug resistance resulted in relapsed and refractory myeloma [5]. Therefore, there is a need to explore possible approaches to enhance the sensitivity of MM to these drugs.
Previous studies discovered that CRBN functions as a substrate receptor of cullin 4-RING E3 ligase (CRL4CRBN) using proteomic and biochemical approaches [6,7,8,9]. CRL4CRBN consists of cullin 4A/B, damage-specific DNA-binding protein 1 (DDB1), RING-box protein ROC1, and CRBN [6, 10]. As a multidomain structural protein, CRBN binds to downstream ubiquitination substrates through its different regions [11]. It has been proposed that CRBN is the primary target of the immunomodulatory drug (IMiD) thalidomide (Thal) and mediates its teratogenic effect [7]. Furthermore, Thal and its structural analogs Len and Pom promoted the ubiquitination-mediated degradation of two essential transcription factors, IKZF1 (Ikaros) and IKZF3 (Aiolos), in MM cells by directly recruiting them to CRBN and thus inhibited the proliferation of MM cells [8, 9]. Given the IMiD-induced recruitment of new substrates (neo-substrates), CRBN has been extensively used to degrade neo-substrates through proteolysis-targeting chimeras (PROTACs) for potential targeted cancer therapy [12,13,14].
Len could block the ubiquitination of CRBN and thus elevate its protein level after short-time treatment [15]. However, long-term Len treatment to myeloma cells caused a decrease in CRBN expression and thus resulted in the Len resistance of myeloma cells [16, 17], suggesting that elevation of CRBN protein levels would enhance the antimyeloma activity of Len [18]. In line with this, arsenic trioxide increased CRBN expression and thus potentiated the antimyeloma effect of Len [19], demonstrating that regulation of CRBN mRNA level could affect its protein level and modulate Len sensitivity. The CSN9 signalosome attenuated the ubiquitination-mediated degradation of CRBN, thereby potentiating the antimyeloma effect of Len [20, 21]. Our previous studies showed that the blockage of caspase-8 increased the protein level of CRBN and thus enhanced the antimyeloma effect of Len [22, 23]. Taken together, these studies indicated that regulation of CRBN at the protein level could affect its stability and modulate Len sensitivity. However, it is largely unknown whether other biological processes such as lysosome-mediated protein degradation could affect CRBN stability and its biological functions.
In this study, we screened the CRBN proximitome and identified BRCC36 as a CRBN-interacting protein using the proximity labeling technique TuboID and quantitative proteomics. We disclosed that BRCC36 removed the K63-linked polyubiquitin chain on CRBN and inhibited its autophagy–lysosome degradation. Furthermore, we demonstrated that the small-molecule compound SHIN1 increased BRCC36 by disrupting its interaction with SHMT2, thereby leading to the upregulation of CRBN and increase of the antimyeloma effect of Len. The discovery in this work revealed a new regulatory mechanism by which CRBN is upregulated and might benefit for the development of therapeutic strategy for the treatment of myeloma patients.
Materials and methods
Reagents
Small-molecule compounds and reagents used in this work were purchased from the following companies: bafilomycin A1 (Baf A1, HY-100558), cycloheximide (CHX, HY-12320), and SHIN1 (HY-112066) were obtained from MedChemExpress. Cell Counting Kit-8 (CCK-8, B34304), lenalidomide (Len, CC-5013), and MG132 (S2619) were ordered from Selleck. NeutrAvidin agarose resin (29,201) was purchased from Thermo Fisher Scientific and biotin (V900418) from Sigma–Aldrich. Protein A/G agarose resin (36403ES08) was acquired from Yeasen Biotechnology (Shanghai, China).
Antibodies were purchased from the following companies: anti-ubiquitin (Ub, sc-8017) from Santa Cruz Biotechnology; anti-CRBN (D8H3S) antibody from Cell Signaling Technology; anti-β-actin (20536–1-AP), anti-β-tubulin (10068–1-AP), anti-Flag (20543–1-AP), and anti-IKZF3 (13561–1-AP) antibodies from Proteintech group; anti-BRCC36 (CQA3926), anti-GFP (CPA9056), anti-HA (CPA9058), anti-RAP80 (CQA3301), and anti-SHMT2 (CQA1041) antibodies from Cohesion Biosciences; anti-IKZF1 (MB0092) antibody from Bioworld. The mouse anti-CRBN antibody was a gift from Dr. Xiu-Bao Chang (Mayo Clinic College of Medicine, USA). The secondary antibodies (111–035-045 and 115–035-062) were from Jackson ImmunoResearch.
Plasmid construction
Plasmids were constructed according to a previous procedure [24,25,26]. 3 × HA-TurboID-NLS-pcDNA3.1 plasmid was from Addgene (107,171). CRBN-Flag-NLS (nuclear localization signal)-TurboID and CRBN-Flag-NES (nuclear export signal)-TurboID plasmids were constructed using polymerase chain reaction (PCR) and ClonExpress Ultra One Step Cloning Kit (C115-02, Vazyme). The shRNA plasmids were constructed using pLKO.1-TRC lentiviral vector according to a previous method [27]. The BRCC36 forward oligonucleotide (5’-CCGGGAGGAAGGACCGAGTAGAAATCTCGAGATTTCTACTCGGTCCTTCCTCTTTTTG-3’) and reverse complementary oligonucleotide (5’-AATTCAAAAAGAGGAAGGACCGAGTAGAAATCTCGAGATTTCTACTCGGTCCTTCCTC-3’) were synthesized by GeneWiz (China). All constructed plasmids were verified by DNA sequencing (GeneWiz).
Cell lines and cell culture
HEK293T, cervical cancer cell line HeLa, and multiple myeloma cell lines MM.1S, RPMI-8226, LP1, U266, and ARH-77 were obtained from American Type Culture Collection (ATCC). MM.1S, RPMI-8226, LP1, U266, and ARH-77 cells were cultured in RPMI 1640 basic medium (C11875500BT, Gibco). HEK293T and HeLa cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM, SH30243.01, HyClone). Growth medium was supplemented with 10% fetal bovine serum (FBS, YS210414, EallBio Life Sciences),100 unit/mL penicillin and 100 µg/mL streptomycin (C100C5, NCM Biotech).
Construction of stable cell lines
Lentiviral particles were made according to a previously described method [28]. Briefly, packaging plasmids psPAX2 and pMD2.G were transfected together with pLKO.1-TRC and pLKO.1-shBRCC36 into HEK293T cells. The myeloma LP1 cells were infected with the lentiviral particles, and then selected with 1 μg/mL puromycin (P8230, Solarbio Life Sciences) for two weeks.
siRNA and plasmid transfection
siRNAs were synthesized by Guangzhou RiboBio Co (China). HEK293T cells were transfected with siNC (Cat #: 160818), siBRCC36 (target sequence: GAGGAAGGACCGAGTAGAA), siSHMT2 (target sequence: CGGAGAGTTGTGGACTTTATA), and siRAP80 (target sequence: CCAGTTGGAGGTTTATCAA) using LipoRNAiMAX (13778–150, Invitrogen) or with plasmids using Lipofectamine 3000 (L3000-015, Invitrogen).
Quantitative PCR (qPCR)
The qPCR was performed according to the previous method [29]. Briefly, total RNAs were isolated using TRIzol (R401-01, Vazyme) and then the HiScript III All-in-one RT SuperMix (R333, Vazyme) was used to synthesize the cDNA library. The Biotool SYBR Green One Step qRT-PCR kit was used to perform the qPCR on the Applied Biosystems 7500 Real-Time PCR system. All results were normalized with GAPDH. The qPCR primers (CRBN forward: ATGCTGAGACCTTAATGGACAGA, CRBN reverse: AAGTCGCTGGATAGCACTGC; BRCC36 forward: GAGTCTGACGCTTTCCTCGTT, BRCC36 reverse: TGTATCATCGTTCAACTCCCCT; GAPDH forward: TGCACCACCAACTGCTTAGC, GAPDH reverse: ACAGTCTTCTGGGTGGCAGTG) were synthesized and purified with HPLC by GeneWiz.
CCK-8 assay
The relative cell viability was measured using CCK-8 assay. Cells were seeded and cultured in 96-well plates (5000 cells/well) for different times after drug treatment. CCK-8 reagent (10 μL/well) was added to the 96-well plates and incubated for 1 h at 37 °C. The absorbance was measured at 450 nm by Tecan Infinite M1000 PRO (Switzerland).
Experiments for protein degradation and proteasome inhibition
The cycloheximide (CHX)-chase experiments were performed to measure the protein degradation. HEK293T cells overexpressing Flag or Flag-BRCC36 were treated with CHX (200 μg/mL) for the indicated times. For proteasome inhibition experiments, HEK293T cells stably expressing shNC or shBRCC36 were treated with DMSO or Baf A1 (100 nM, 24 h). The cell lysates were harvested and subjected to immunoblotting analysis.
Xenograft mouse model
The Male BALB/c nude mice (3–5 week old,16-18 g) were purchased from the Shanghai SLRC Laboratory Animal Co. The LP1 cells were injected subcutaneously into the right flank of the nude mice. The mice were randomly divided into four groups (n = 3) when tumor size reached 100 mm3 after injection. Control, SHIN1, Len, SHIN1 and Len were then administered by intraperitoneal injection each other day for three times. The length (a) and width (b) of the tumor were measured each other day for two weeks and the tumor size was calculated by a × b2/2. The tumor tissues were dissected and weighted for further western blotting analysis. All animal work was approved by the Animal Ethics Committee of Soochow University.
Western blotting analysis
Cell lysates or immunoprecipitates were harvested and subjected to Western blotting according to a previously described method [30]. The NcmECL Ultra substrate (P10300, NCM Biotech) was used to visualize and analyze the protein expression.
Affinity purification
Flag tagged proteins were purified through a method describe previously [31]. HEK293T cell lysates were incubated with anti-Flag affinity gel (B23102, Bimake) at 4 °C overnight. The gel was then washed three times with TBST (TBS with 0.1% Tween 20) and modified RIPA buffer. Flag-tagged proteins were eluted with a solution containing the Flag peptide (200 μg/mL, B23112, Bimake). Biotinylated proteins were purified with NeutrAvidin agarose resin based on a previous study [32]. The samples were analyzed through Western blotting according to a previous procedure [31, 33].
Proximity-labeling and MS analysis
Proximity-dependent biotinylation has been optimized in our previous study [34]. Basically, HEK293T cells were transfected with pcDNA3.1, CRBN-Flag-NLS-TurboID, and CRBN-Flag-NES-TurboID for 48 h, and then treated with biotin (50 μM) for 30 min. The biotinylated proteins from cell lysates were digested for MS analysis using a procedure described previously [34].
Results
Proximity labeling technique TurboID and quantitative proteomics identify the deubiquitinase BRCC36 as a CRBN-interacting protein
Generally, proteins execute their biological functions through their interacting proteins. To systematically identify CRBN-interacting proteins, we employed the proximity labeling technique TurboID and screened the CRBN proximitome by quantitative proteomics (Fig. 1A). The previous study showed that subcellular localized CRBN executed different functions in the substrates degradation [35]. To investigate the possible proteins proximal to CRBN in cellular nucleus and cytoplasm, we constructed plasmids expressing nuclear localized CRBN (CRBN-Flag-NLS-TurboID) and cytoplasmic localized CRBN (CRBN-Flag-NES-TurboID) and then transfected these plasmids, respectively, into HEK293 cells (Fig. 1B). The combination of TurboID and quantitative proteomics revealed hundreds of proteins proximal to CRBN (Fig. 1C). We discovered that the Lys-63-specific deubiquitinase BRCC36 was one of the statistically significant CRBN-interacting proteins (Fig. 1D for the identified peptides and Fig. 1E for the MS/MS spectrum of a representative peptide).
Identification of CRBN proximitome by proximity labeling and quantitative proteomics. A Principle of efficient proximity labeling with TurboID. TurboID is fused to CRBN and catalyzes the biotinylation of proteins proximal to CRBN, which were then purified with NeutrAvidin, digested with trypsin, and analyzed by LC–MS/MS. B Immunofluorescence images for HEK293 cells expressing CRBN-Flag-NLS-TurboID and CRBN-Flag-NES-TurboID. Scale bar: 20 μm. C Volcano plot of − Log10 (P-value) versus Log2 (NLS/NES ratio) of proteins identified by MS from CRBN-Flag-NLS-TurboID- and CRBN-Flag-NES-TurboID-expressing cells after NeutrAvidin purification. The P value was calculated using Student’s t-test with data from three biological replicates. *: P < 0.05. D Information for MS-identified tryptic peptides derived from BRCC36. E MS/MS spectrum of a representative tryptic peptide derived from BRCC36
BRCC36 interacts with CRBN
We next sought to validate the interaction between BRCC36 and CRBN using biochemical methods. First, we constructed plasmids expressing BRCC36 and CRBN and then investigated their subcellular distribution. This experiment revealed that BRCC36 is a whole cell distributed protein (Fig. 2A). We then overexpressed CRBN and BRCC36 in HEK293T cells and affinity-purified BRCC36 using anti-Flag affinity beads. We discovered that CRBN was co-immunoprecipitated with BRCC36 but not with the negative control, indicating that exogenous BRCC36 interacts with exogenous CRBN (Fig. 2B). Furthermore, co-immunoprecipitation experiment disclosed that BRCC36 and CRBN interacted with each other endogenously in HEK293T cells and myeloma LP1 cells (Figs. 2C-D). CRBN contained Lon-protease like domian (81–317 AA) and thalidomide binding domain (339–442 AA). To further narrow down the precise binding regions between BRCC36 and CRBN, we constructed several plasmids expressing truncated CRBNs (Fig. 2E). Co-immunoprecipitation unveiled that the 1–81 amino acids in CRBN were not required for the interaction between BRCC36 and CRBN (Fig. 2F), which suggested that the Lon-protease like domain might mediated the interaction between CRBN and BRCC36. Taken together, these data suggested that BRCC36 could indeed bind to CRBN.
CRBN interacts with BRCC36. A CRBN colocalizes with BRCC36 in the cytoplasm and nucleus. HEK293 cells were transfected with the indicated plasmids for 24 h and incubated with primary and secondary fluorescence antibodies. Immunofluorescence was detected by inverted IX71 microscopy system. Scale bar: 20 μm. B CRBN interacts with BRCC36 exogenously. HA-CRBN was expressed with Flag or Flag-BRCC36 in HEK293T cells. After 48 h, cells were lysed and BRCC36 was immunoprecipitated with anti-Flag affinity gel. The immunoprecipitates and cell lysates were subjected to immunoblotting analysis. C, D BRCC36 interacts with CRBN endogenously. HEK293T (C) or LP1 (D) cell lysates were incubated with anti-CRBN antibody or IgG overnight and then incubated with protein A/G agarose resin for 3 h. The immunoprecipitates and cell lysates were subjected to immunoblotting analysis. E The schematic representation of CRBN and its truncations. F The CRBN 82–186 domain is critical for its interaction with BRCC36. HEK293T cells were transfected with Flag-BRCC36 and GFP, GFP-CRBN wild type (WT) or truncation mutants for 48 h. Flag-BRCC36 was purified with anti-Flag affinity gel. Cell lysates and purified samples were immunoblotted for the indicated antibodies
BRCC36 increases CRBN protein level
Since BRCC36 is a deubiquitinase, which could remove K63-linked polyubiquitin chains from substrates, we then investigated whether BRCC36 affected CRBN protein levels. To do so, we transfected Flag-BRCC36 into HEK293T cells and immunoblotted CRBN. The results showed that BRCC36 increased CRBN (Fig. 3A). We further gradually increased the quantity of Flag-BRCC36 plasmid transfected into HEK293T cells and immunoblotting of cell lysates revealed that CRBN was increased progressively (Fig. 3B). Taken together, these data suggested that overexpression of BRCC36 increased CRBN in HEK293T cells. To further confirm these results, we knocked down BRCC36 in HEK293T and HeLa cells and found that CRBN was decreased in these two cell lines (Figs. 3C-D). Similar results were observed when BRCC36 was knocked down in two myeloma cell lines, RPMI-8226 and LP1 (Figs. 3E-F). Taken together, these data suggested that BRCC36 could increase CRBN protein level. In line with this conclusion, myeloma cells expressing high BRCC36 have more CRBN protein level (Supplemental Fig. 1), and the positively correlation between BRCC36 and CRBN in cancer proteomics databases (Supplemental Fig. 2).
BRCC36 stabilizes CRBN. A, B BRCC36 overexpression increased CRBN protein levels. HEK293T cells were transfected with pcDNA3.1 or Flag-BRCC36 plasmid (0, 1, 2, or 4 μg) for 72 h. Cell lysates were analyzed by Western blotting with the indicated antibodies. The mean and standard deviation (Mean ± SD) were depicted, and the P value was calculated using Student’s t-test with data from three biological replicates. *: P < 0.05; **: P < 0.01. C, D Knockdown of BRCC36 decreased CRBN protein levels in HEK293T (C) and HeLa (D) cells. HEK293T and HeLa cells were transfected with siNC and siBRCC36 for 48 h. Cell lysates were analyzed by Western blotting with the indicated antibodies. Experiments were repeated three times for quantification, and Student’s t-test was used to calculate the P values. **: P < 0.01; ***: P < 0.001. E, F Knockdown of BRCC36 decreased CRBN protein levels in MM cell lines. RPMI-8226 (E) and LP1 (F) cells were infected with shNC or shBRCC36 lentiviruses for 48 h. Experiments were repeated three times for quantification, and Student’s t-test was used to calculate the P values. *: P < 0.05; **: P < 0.01
BRCC36 attenuates the K63-ubiquitination-mediated CRBN degradation
After having discovered the upregulation of CRBN by BRCC36, we sought to investigate the underlying molecular mechanism. CHX-chase and qPCR experiments revealed that BRCC36 increased the protein stability of CRBN (Fig. 4A), but not its mRNA level (Fig. 4B), which suggested that BRCC36 might regulate CRBN at the post-transcriptional modification level. It has been reported that the Lys-63-specific deubiquitinase BRCC36 cleaves the K63-linked polyubiquitin chain and thus regulates the DNA damage responses [36]. Consistent with this, we found that the inactive BRCC36 QSQ mutant did not increase CRBN protein level (Fig. 4C). Blockage of the autophagy-lysosome pathway (ALP) using the lysosome hydrogen-ion ATPase inhibitor Baf A1 resulted in the CRBN and BRCC36 increase (Fig. 4D and Supplemental Fig. 3), suggesting that CRBN and BRCC36 could also be degraded by ALP. Furthermore, the regulation of CRBN by BRCC36 could be attenuated by Baf A1 (Fig. 4E), demonstrating that BRCC36 regulated CRBN protein levels through ALP. Because K63 ubiquitination is closely associated with ALP, we then examined the regulation of BRCC36 on the K63 ubiquitination of CRBN. We transfected the wild-type, K48R, or K63R HA-Ub, respectively, into mock and BRCC36-deficient HEK293T cells. Immunoblotting of affinity-purified CRBN showed that CRBN ubiquitination was elevated when the wild-type or K48R Ub but not the K63R Ub was expressed after knocking down BRCC36, which indicated that BRCC36 could diminish the K63 ubiquitination on CRBN (Fig. 4F).
BRCC36 protected CRBN from degradation by autophagy–lysosome pathway. A CHX chase experiments revealed that overexpression of BRCC36 diminished the degradation of CRBN. HEK293T cells were expressed with Flag-BRCC36 for 48 h and then treated with DMSO or CHX (200 μg/mL) for the indicated times. Cell lysates were immunoblotted and the relative protein level was quantified. Experiments were repeated three times for quantification. Two-way ANOVA was used to calculate the P values. **: P < 0.01. B BRCC36 knockdown did not decrease CRBN mRNA levels. HEK293T cells were transfected with siNC and siBRCC36 for 48 h. Total RNA was extracted, cDNA was reverse-transcribed, and mRNA was analyzed by RT–qPCR. Experiments were repeated three times for quantification. Student’s t-test was used to calculate the P values. ns not significant. C The regulation of CRBN by BRCC36 depended on its enzymatic activity. HEK293T cells were transfected with Flag, Flag-BRCC36, or Flag-BRCC36-QSQ (Flag-QSQ) for 72 h. Cell lysates were analyzed by Western blotting with the indicated antibodies. Experiments were repeated three times for quantification. Student’s t-test was used to calculate the P values. **: P < 0.01; ns not significant. D CRBN was degraded through ALP. HEK293T cells were transfected with HA-CRBN and treated with DMSO or bafilomycin A1 (Baf A1) for 24 h. Cell lysates were analyzed by Western blotting with the indicated antibodies. Experiments were repeated three times for quantification. Student’s t-test was used to calculate the P values. ***: P < 0.001. E Baf A1 attenuated the BRCC36-induced CRBN increase. HEK293T cells were transfected with HA-CRBN and Flag-BRCC36 or Flag for 48 h and then treated with DMSO or Baf A1 for 24 h. Cell lysates were immunoblotted with the indicated antibodies. F BRCC36 downregulated the K63-linked polyubiquitination on CRBN. HEK293T cells were transfected with the indicated plasmids for 48 h and purified with anti-Flag affinity gel. The immunoprecipitates and cell lysates were subjected to immunoblotting analysis
Small-molecule compound SHIN1 increases CRBN through disrupting the BRISC complex
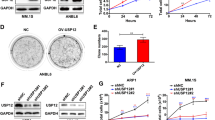
It has been reported that BRCC36 exerts enzyme activity through two different protein complexes BRCA1-A and BRISC [37]. We then knocked down the two essential components of these complexes, RAP80 and SHMT2, respectively, and then investigated the subsequent effect on CRBN. The immunoblotting images indicated that CRBN and BRCC36 were decreased in SHMT2-deficient cells, but not in RAP80-deficient cells (Fig. 5A-5B). SHIN1 is an SHMT2 inhibitor that can disrupt the SHMT2-BRCC36 complex (Supplemental Fig. 4). We then treated HEK293T and HeLa cells with SHIN1 and examined the protein level of CRBN and found that SHIN1 could significantly increase CRBN in these two cell lines (Fig. 5C). To further confirm this result, we treated the myeloma cell lines LP1, RPMI-8226, ARH-77, and U266 with SHIN1 and discovered that SHIN1 could also increase CRBN in these myeloma cell lines (Fig. 5D). Furthermore, we failed to detect the increase of CRBN in BRCC36-deficient cells upon SHIN1 treatment, suggesting that SHIN1 increased CRBN in a BRCC36-dependent manner (Fig. 5E). Given the function of ALP on the regulation of CRBN, we further investigated the possible effect of SHIN1 on ALP. However, we could not observe the influence of SHIN1 on ALP (Supplemental Fig. 5).
BRISC complex mediates the regulation of CRBN by BRCC36. A, B Knockdown of SHMT2 (A) but not RAP80 (B) reduced CRBN. HEK293T cells were transfected with siNC and siSHMT2 or siRAP80 for 48 h. Cell lysates were immunoblotted with the indicated antibodies. Experiments were repeated three times for quantification. Student’s t-test was used to calculate the P values. **: P < 0.01; ns: not significant. C, D SHIN1 upregulated CRBN protein levels in HeLa, HEK293T (C), and myeloma cell lines (D). HeLa, HEK293T, LP1, U266, RPMI-8226, and ARH-77 cells were treated with SHIN1 (10 μM) or DMSO for 24 h. Cell lysates were analyzed by Western blotting with the indicated antibodies. Experiments were repeated three times for quantification. Student’s t-test was used to calculate the P values. *: P < 0.05; **: P < 0.01. E BRCC36 was required for the upregulation of CRBN by SHIN1. HeLa cells stably expressing shNC and shBRCC36 were treated with DMSO or SHIN1 (10 μM) for 24 h. Cell lysates were immunoblotted with the indicated antibodies. Experiments were repeated three times for quantification. Student’s t-test was used to calculate the P values. *: P < 0.05; **: P < 0.01; ns not significant
SHIN1 enhances the antimyeloma effect of Len in myeloma cell lines
Given that SHIN1 increased CRBN, which mediates the Len-induced inhibitory effect on the proliferation of myeloma cells, we further explored whether SHIN1 could modulate the antimyeloma activity of Len in myeloma cells. We treated myeloma cell lines MM1.S, LP1, and RPMI-8226 with SHIN1 and/or Len. CCK-8 assays demonstrated that SHIN1 enhanced the antimyeloma effect of Len (Figs. 6A-C). We also discovered that SHIN1 could improve the antimyeloma effect of Len at a lower dose (Supplemental Fig. 6). The biochemical experiments showed that SHIN1 could further enhance the Len-regulated decrease of IKZF1 and IKZF3 in myeloma cell lines (Figs. 6A-C). SHIN1 could enhance the antimyeloma effect of Len and decrease IKZF1 and IKZF3 in a dose-dependent manner (Supplemental Fig. 7 and 8A). However, the combination of SHIN1 and Len did not induce the apoptosis in LP1 cells (Supplemental Fig. 8B). Furthermore, Long-term Len treatment for myeloma cells would decrease CRNB at the protein and mRNA level, and SHIN1 did not regulate the antimyeloma effect of Len in these Len-resistant cells (Supplemental Fig. 9). Taken together, these data suggested that the small-molecule compound SHIN1 could potentiate the antimyeloma effect of Len by upregulating CRBN.
SHIN1 enhances the antimyeloma activity of Len in myeloma cells. A–C Left panels: MM.1S (A), LP1 (B), and RPMI-8226 (C) cells were pretreated with DMSO or SHIN1 (100 nM) for 24 h and then treated with DMSO or Len (10 μM) for 5 days. The relative cell viability was determined by CCK-8 assay. Experiments were repeated three times for quantification, and Student’s t-test was used to calculate the P values. *: P < 0.05; **: P < 0.01; ***: P < 0.001 (left panels). Right panels: MM.1S, LP1, and RPMI-8226 cells were pretreated with DMSO or SHIN1 (100 nM) for 24 h and then treated with DMSO or Len (10 μM) for 3 h. Cell lysates were immunoblotted with the indicated antibodies
SHIN1 enhances the antimyeloma effect of Len in xenograft mice
We further explored whether SHIN1 could modulate the antimyeloma activity of Len in vivo. We treated xenograft mice inoculating LP1 cells with SHIN1 and/or Len. The combination of SHIN1 and Len could markedly decrease the LP1 tumor growth rate (Fig. 7A-D), which demonstrated that SHIN1 enhanced the antimyeloma effect of Len in vivo. We also analyzed the tumor tissues from the xenografts, and discovered that SHIN1 could decrase IKZF1 and IKZF3 in the presence of Len (Fig. 7E). Taken together, these data suggested that the small-molecule compound SHIN1 could potentiate the antimyeloma effect of Len in vitro and in vivo (Fig. 7F).
SHIN1 enhances the antimyeloma activity of Len in xenograft mice. A–D LP1 cells were injected into SCID mice subcutaneously. At 14 days after transplantation, control (10% DMSO, 40% PEG300, 5% Tween-80, and 45% Saline), SHIN1 (50 mg/kg/d), and/ or Len (50 mg/kg/d) was administered to the mice (n = 3) by intraperitoneal injection every other day for three times. The length and width of the tumors were measured every other day for two weeks. Tumor weights, body weights, and tumor volumes were then measured at the end of the experiments. Student’s t-test and two-way ANOVA were used to calculate the P values. *: P < 0.05; **: P < 0.01; ***: P < 0.001; ****: P < 0.0001. E Cell lysates were prepared from tumor tissues of the xenograft mice for immunoblotting of the indicated proteins. Experiments were repeated three times from three different mice for quantification, and Student’s t-test was used to calculate the P values. *: P < 0.05; **: P < 0.01; ***: P < 0.001. F Proposed model for the regulation of BRCC36 on the antimyeloma effect of Len. The CRBN-interacting protein deubiquitinase BRCC36 can cleave the K63-linked polyubiquitin chains on CRBN and protect CRBN from lysosomal degradation. SHIN1, the small-molecule inhibitor of SHMT2, disrupts the BRISC complex and reduces CRBN degradation, and thus enhances the sensitivity of multiple myeloma cells to IMiDs
Discussion
In this study, by employing TurboID and quantitative proteomics, we identified a new CRBN-interacting protein BRCC36, a Lys-63-specific deubiquitinase. We further demonstrated that BRCC36 interacted with CRBN, stabilized CRBN protein, and protected CRBN from lysosomal degradation by removing the K63-linked polyubiquitin chains from CRBN. This result is consistent with a previous work, which disclosed that BRCC36 cleaved the K63-linked polyubiquitin chains on IFNAR1, thus limiting its lysosomal degradation and thereby increasing its protein stability and activation [38]. Furthermore, we disclosed that the SHMT2 inhibitor SHIN1 could increase CRBN protein level through disrupting SHMT2-BRCC36 complex and thus potentiate the antimyeloma effect of Len, which provides a possible new therapeutic strategy for myeloma.
CRBN is a substrate receptor of the CRL4 E3 ligase and mediates the degradation of neo-substrates IKZF1/IKZF3 upon IMiDs treatment in myeloma cells, which eventually inhibits the proliferation of myeloma cells [8, 9]. However, long-term and IMiD therapy resulted in relapse and resistance in myeloma. Discovering new combination therapy would benefit myeloma patients. Long-term Len treatment results in the loss of CRBN at the protein and mRNA levels [16, 18], suggesting that elevation of CRBN protein levels potentiates the antimyeloma effect of Len [17, 39]. In line with this, attenuation of CRBN ubiquitination-proteasome degradation and its cleavage could increase CRBN protein level and thus potentiates the antimyeloma effect of Len [21,22,23].
It has been revealed that CRBN is degraded through the ubiquitin–proteasome system (UPS) [15, 39]. Consistent with these previous studies, we confirmed that CRBN was increased upon the blockade of the proteasome. The C-terminal truncation mutation of CRBN caused mild degree of intellectual disability in children, suggesting the crucial role of CRBN in neurological diseases [40]. A missense mutation of CRBN also resulted in severe degree of intellectual disability, which further supported the above conclusion [41], although the underlying molecular mechanisms were waiting to be uncovered. Proteasomal inhibition could recover the protein level of C-terminal truncated CRBN, demonstrating that the C-terminus of CRBN enhanced the protein stability [15]. The proteomic data published previously proposed that there were multiple ubiquitination sites on CRBN, which affect its stability [15]. Here, we further proposed that CRBN could also be modified by K63-linked polyubiquitin chains and thus undergo degradation through ALP, although the precise sites for K63 ubiquitination are yet known.
The proteasome inhibitor bortezomib (PS-341) was approved by the FDA for the treatment of myeloma in 2003 [42] and has improved the treatment of multiple myeloma in recent decades. The combination treatment of bortezomib and Len for myeloma patients has been the first-line therapy [43], although there are paradoxical pharmacological mechanisms [44]. Furthermore, bortezomib induced CRBN cleavage in myeloma cells through activating caspase-8, suggesting that the effect of combination of bortezomib and Len treatment for myeloma would be attenuated in patients with high caspase-8 expression [22, 23]. Here, we found one alternative solution to elevate CRBN protein levels through reducing its degradation via ALP but not UPS. Furthermore, the addition of SHIN1 increased CRBN protein level through enhancing the deubiquitinating activity of BRCC36.
It should be noted that the BRISC complex regulates immune signaling in response to inflammatory stimuli [45]. The elevation of CRBN might decrease the constitutive substrate c-Jun and then suppress the immune response [46]. In our study, the regulation of SHIN1-BRCC36 on CRBN was observed in both myeloma and non-myeloma cell lines, suggesting that SHIN1-BRCC36 might modulate the immune response in the tumor microenvironment and thus regulate the antimyeloma effect of Len.
Given the higher efficiency of the UPS on the degradation of short-lived substrates than the ALP, CRBN most probably is degraded mainly through the UPS. Therefore, the effect of ALP on the stability of CRBN would be limited, although it is significantly increased upon the blockage of ALP in this study (Fig. 4D). Furthermore, the substrates were degraded through the UPS in a highly specific fashion, which depends on the enzymes involved in protein ubiquitination and the topology of the ubiquitin chains. However, the ALP is less targeted than the UPS. Therefore, the ALP inhibitor would generate multiple side effects although it might benefit the Len-based therapy in multiple myeloma. Nevertheless, this work revealed that CRBN undergoes lysosomal degradation and SHIN1-BRCC36 axis reduces this degradation, thus potentiating the antimyeloma effect of Len, which may provide an alternative combination therapy for myeloma patients.
Data availability
The MS data were deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProx partner repository with the data set identifier PXD045100 (https://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD045100 or https://www.iprox.cn//page/project.html?id=IPX0007056000).
References
Kazandjian D (2016) Multiple myeloma epidemiology and survival: A unique malignancy. Semin Oncol 43(6):676–681
Goldschmidt H, Ashcroft J, Szabo Z, Garderet L (2019) Navigating the treatment landscape in multiple myeloma: which combinations to use and when? Ann Hematol 98(1):1–18
van de Donk N, Usmani SZ (2018) CD38 antibodies in multiple myeloma: mechanisms of action and modes of resistance. Front Immunol 9:2134
Lee JH, Kim SH (2020) Treatment of relapsed and refractory multiple myeloma. Blood Res 55(S1):S43–S53
Hernandez-Rivas JA, Rios-Tamayo R, Encinas C, Alonso R, Lahuerta JJ (2022) The changing landscape of relapsed and/or refractory multiple myeloma (MM): fundamentals and controversies. Biomark Res 10(1):1
Angers S, Li T, Yi X, MacCoss MJ, Moon RT, Zheng N (2006) Molecular architecture and assembly of the DDB1–CUL4A ubiquitin ligase machinery. Nature 443(7111):590–593
Ito T, Ando H, Suzuki T, Ogura T, Hotta K, Imamura Y et al (2010) Identification of a primary target of thalidomide teratogenicity. Science 327(5971):1345–1350
Lu G, Middleton RE, Sun H, Naniong M, Ott CJ, Mitsiades CS et al (2014) The myeloma drug lenalidomide promotes the cereblon-dependent destruction of Ikaros proteins. Science 343(6168):305–309
Kronke J, Udeshi ND, Narla A, Grauman P, Hurst SN, McConkey M et al (2014) Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science 343(6168):301–305
Jackson S, Xiong Y (2009) CRL4s: the CUL4-RING E3 ubiquitin ligases. Trends Biochem Sci 34(11):562–570
Tao J, Yang J, Xu G (2018) The interacting domains in cereblon differentially modulate the immunomodulatory drug-mediated ubiquitination and degradation of its binding partners. Biochem Biophys Res Commun 507(1–4):443–449
Wang Y, Jiang X, Feng F, Liu W, Sun H (2020) Degradation of proteins by PROTACs and other strategies. Acta Pharm Sin B 10(2):207–238
Winter GE, Buckley DL, Paulk J, Roberts JM, Souza A, Dhe-Paganon S et al (2015) Drug Development. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science 348(6241):1376–1381
Lu J, Qian Y, Altieri M, Dong H, Wang J, Raina K et al (2015) Hijacking the E3 ubiquitin ligase cereblon to efficiently target BRD4. Chem Biol 22(6):755–763
Xu G, Jiang X, Jaffrey SR (2013) A mental retardation-linked nonsense mutation in cereblon is rescued by proteasome inhibition. J Biol Chem 288(41):29573–29585
Lopez-Girona A, Mendy D, Ito T, Miller K, Gandhi AK, Kang J et al (2012) Cereblon is a direct protein target for immunomodulatory and antiproliferative activities of lenalidomide and pomalidomide. Leukemia 26(11):2326–2335
Gandhi AK, Mendy D, Waldman M, Chen G, Rychak E, Miller K et al (2014) Measuring cereblon as a biomarker of response or resistance to lenalidomide and pomalidomide requires use of standardized reagents and understanding of gene complexity. Br J Haematol 164(2):233–244
Zhu YX, Braggio E, Shi CX, Bruins LA, Schmidt JE, Van Wier S et al (2011) Cereblon expression is required for the antimyeloma activity of lenalidomide and pomalidomide. Blood 118(18):4771–4779
Jian Y, Gao W, Geng C, Zhou H, Leng Y, Li Y et al (2017) Arsenic trioxide potentiates sensitivity of multiple myeloma cells to lenalidomide by upregulating cereblon expression levels. Oncol Lett 14(3):3243–3248
Sievers QL, Gasser JA, Cowley GS, Fischer ES, Ebert BL (2018) Genome-wide screen identifies cullin-RING ligase machinery required for lenalidomide-dependent CRL4CRBN activity. Blood 132(12):1293–1303
Liu J, Song T, Zhou W, Xing L, Wang S, Ho M et al (2019) A genome-scale CRISPR-Cas9 screening in myeloma cells identifies regulators of immunomodulatory drug sensitivity. Leukemia 33(1):171–180
Zhou L, Yu W, Jayabalan DS, Niesvizky R, Jaffrey SR, Huang X et al (2020) Caspase-8 inhibition prevents the cleavage and degradation of E3 ligase substrate receptor cereblon and potentiates its biological function. Front Cell Dev Biol 8(1562):605989
Zhou L, Huang X, Niesvizky R, Pu Z, Xu G (2021) Caspase-8 regulates the antimyeloma activity of bortezomib and lenalidomide. J Pharmacol Exp Ther 379(3):303–309
Duan Q, Li D, Xiong L, Chang Z, Xu G (2019) SILAC quantitative proteomics and biochemical analyses reveal a novel molecular mechanism by which ADAM12S promotes the proliferation, migration, and invasion of small cell lung cancer cells through upregulating hexokinase 1. J Proteome Res 18(7):2903–2914
Hu Z, Wang X, Li D, Cao L, Cui H, Xu G (2021) UFBP1, a key component in ufmylation, enhances drug sensitivity by promoting proteasomal degradation of oxidative stress-response transcription factor Nrf2. Oncogene 40(3):647–662
Chen D, Li Y-P, Yu Y-X, Zhou T, Liu C, Fei E-K et al (2018) Dendritic cell nuclear protein-1 regulates melatonin biosynthesis by binding to BMAL1 and inhibiting the transcription of N-acetyltransferase in C6 cells. Acta Pharmacol Sin 39(4):597–606
Liu G, Liu Q, Yan B, Zhu Z, Xu Y (2020) USP7 inhibition alleviates H2O2-induced injury in chondrocytes via inhibiting NOX4/NLRP3 pathway. Front Pharmacol 11(2480):617270
Zhang Q-Q, Wang W-j, Li J, Yang N, Chen G, Wang Z et al (2015) Cathepsin L suppression increases the radiosensitivity of human glioma U251 cells via G2/M cell cycle arrest and DNA damage. Acta Pharmacol Sin 36(9):1113–1125
Chen XH, Xu YJ, Wang XG, Lin P, Cao BY, Zeng YY et al (2019) Mebendazole elicits potent antimyeloma activity by inhibiting the USP5/c-Maf axis. Acta Pharmacol Sin 40(12):1568–1577
Hou X, Si J, Ren H, Chen D, Wang H, Ying Z et al (2015) Parkin represses 6-hydroxydopamine-induced apoptosis via stabilizing scaffold protein p62 in PC12 cells. Acta Pharmacol Sin 36(11):1300–1307
Yu W, Wang B, Zhou L, Xu G (2021) Endoplasmic reticulum stress-mediated p62 downregulation inhibits apoptosis via c-Jun upregulation. Biomol Ther (Seoul) 29(2):195–204
Lu J, Jiang H, Li D, Chen T, Wang Y, Pu Z et al (2021) Proximity labeling, quantitative proteomics, and biochemical studies revealed the molecular mechanism for the inhibitory effect of indisulam on the proliferation of gastric cancer cells. J Proteome Res 20(9):4462–4474
Guo DK, Zhu Y, Sun HY, Xu XY, Zhang S, Hao ZB et al (2019) Pharmacological activation of REV-ERBα represses LPS-induced microglial activation through the NF-µB pathway. Acta Pharmacol Sin 40(1):26–34
Wang X, Cao L, Jiang H, Zhou L, Hu Z, Xu G (2023) Proximity proteomics and biochemical analysis reveal a noncanonical function for UFM1-specific protease 1 in the p62 body formation. J Proteome Res 22(7):2352–2363
Tateno S, Iida M, Fujii S, Suwa T, Katayama M, Tokuyama H et al (2020) Genome-wide screening reveals a role for subcellular localization of CRBN in the anti-myeloma activity of pomalidomide. Sci Rep 10(1):4012
Ng HM, Wei L, Lan L, Huen MS (2016) The Lys63-deubiquitylating enzyme BRCC36 Limits DNA break processing and repair. J Biol Chem 291(31):16197–16207
Rabl J (2020) BRCA1-A and BRISC: multifunctional molecular machines for ubiquitin signaling. Biomolecules 10(11):1503
Zheng H, Gupta V, Patterson-Fortin J, Bhattacharya S, Katlinski K, Wu J et al (2013) A BRISC-SHMT complex deubiquitinates IFNAR1 and regulates interferon responses. Cell Rep 5(1):180–193
Liu Y, Huang X, He X, Zhou Y, Jiang X, Chen-Kiang S et al (2015) A novel effect of thalidomide and its analogs: suppression of cereblon ubiquitination enhances ubiquitin ligase function. FASEB J 29(12):4829–4839
Higgins JJ, Pucilowska J, Lombardi RQ, Rooney JP (2004) A mutation in a novel ATP-dependent Lon protease gene in a kindred with mild mental retardation. Neurology 63(10):1927–1931
Sheereen A, Alaamery M, Bawazeer S, Al Yafee Y, Massadeh S, Eyaid W (2017) A missense mutation in the CRBN gene that segregates with intellectual disability and self-mutilating behaviour in a consanguineous Saudi family. J Med Genet 54(4):236–240
Kane RC, Bross PF, Farrell AT, Pazdur R (2003) Velcade: U.S. FDA approval for the treatment of multiple myeloma progressing on prior therapy. Oncologist 8(6):508–513
Zou Y, Lin M, Sheng Z, Niu S (2014) Bortezomib and lenalidomide as front-line therapy for multiple myeloma. Leuk Lymphoma 55(9):2024–2031
Yang J, Zhou Y, Xie S, Wang J, Li Z, Chen L et al (2021) Metformin induces Ferroptosis by inhibiting UFMylation of SLC7A11 in breast cancer. J Exp Clin Cancer Res 40(1):206
Walden M, Tian L, Ross RL, Sykora UM, Byrne DP, Hesketh EL et al (2019) Metabolic control of BRISC–SHMT2 assembly regulates immune signalling. Nature 570(7760):194–199
Yang J, Huang M, Zhou L, He X, Jiang X, Zhang Y et al (2018) Cereblon suppresses the lipopolysaccharide-induced inflammatory response by promoting the ubiquitination and degradation of c-Jun. J Biol Chem 293(26):10141–10157
Acknowledgements
The anti-CRBN mouse antibody was a kind gift from Dr. Xiu-Bao Chang at Mayo Clinic College of Medicine (USA).
Funding
This work was supported by National Natural Science Foundation of China (32170975 & 32171437), the Suzhou Science and Technology Project (SKY2022115), the Natural Science Foundation of the Jiangsu Higher Education Institutions of China (22KJB310017), Gusu Key Health Talent Program of Suzhou (GSWS2022122), Jiangsu Key Laboratory of Neuropsychiatric Diseases (BM2013003), National Center for International Research (2017B01012), and a project funded by the Postgraduate Research & Practice Innovation Program of Jiangsu Province.
Author information
Authors and Affiliations
Contributions
LZ, HR, JM, and GX designed the research. BW, ML, DC, QS and WY performed the research. BW, ML, DC, QS, WY, LZ, HR, and GX analyzed the data. BW, JM, LZ, HR, and GX wrote, revised, and reviewed the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Ethical approval and consent to participate
This study did not involve research of human subject, and did not require the ethics approval.
Consent for publication
No consent to publish had to be obtained.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wang, B., Li, M., Cao, D. et al. Lys-63-specific deubiquitinase BRCC36 enhances the sensitivity of multiple myeloma cells to lenalidomide by inhibiting lysosomal degradation of cereblon. Cell. Mol. Life Sci. 81, 349 (2024). https://doi.org/10.1007/s00018-024-05390-1
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-024-05390-1