Abstract
Background
Oxidative stress induced growth inhibitor 1 (OSGIN1) regulates cell death. The role and underlying molecular mechanism of OSGIN1 in non-small cell lung cancer (NSCLC) are uncharacterized.
Methods
OSGIN1 expression in NSCLC samples was detected using immunohistochemistry and Western blotting. Growth of NSCLC cells and gefitinib-resistant cells expressing OSGIN1 or TUBB3 knockdown was determined by MTT, soft agar, and foci formation assays. The effect of OSGIN1 knockdown on in vivo tumor growth was assessed using NSCLC patient-derived xenograft models and gefitinib-resistant patient-derived xenograft models. Potentially interacting protein partners of OSGIN1 were identified using IP-MS/MS, immunoprecipitation, PLA, and Western blotting assays. Microtubule dynamics were explored by tubulin polymerization assay and immunofluorescence. Differential expression of signaling molecules in OSGIN1 knockdown cells was investigated using phospho-proteomics, KEGG analysis, and Western blotting.
Results
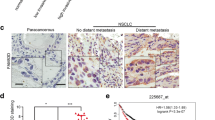
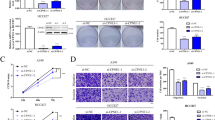
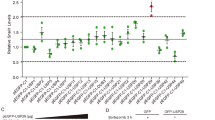
We found that OSGIN1 is highly expressed in NSCLC tissues and is positively correlated with low survival rates and tumor size in lung cancer patients. OSGIN1 knockdown inhibited NSCLC cell growth and patient-derived NSCLC tumor growth in vivo. Knockdown of OSGIN1 strongly increased tubulin polymerization and re-established gefitinib sensitivity in vitro and in vivo. Additionally, knockdown of TUBB3 strongly inhibited NSCLC cell proliferation. Mechanistically, we found that OSGIN1 enhances DYRK1A-mediated TUBB3 phosphorylation, which is critical for inducing tubulin depolymerization. The results of phospho-proteomics and ontology analysis indicated that knockdown of OSGIN1 led to reduced propagation of the MKK3/6-p38 signaling axis.
Conclusions
We propose that OSGIN1 modulates microtubule dynamics by enhancing DYRK1A-mediated phosphorylation of TUBB3 at serine 172. Moreover, elevated OSGIN1 expression promotes NSCLC tumor growth and gefitinib resistance through the MKK3/6-p38 signaling pathway. Our findings unveil a new mechanism of OSGIN1 and provide a promising therapeutic target for NSCLC treatment in the clinic.








Similar content being viewed by others
Availability of data and materials
The data are available to academic researchers upon request.
Abbreviations
- NSCLC:
-
Non-small cell lung cancer
- OSGIN1:
-
Oxidative stress induced growth inhibitor 1
- TUBB3:
-
Tubulin beta 3 class III
- DYRK1A:
-
Dual specificity tyrosine phosphorylation regulated kinase 1A
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- MTT:
-
Methyl thiazolyl tetrazolium
- IP:
-
Immunoprecipitation
- MS:
-
Mass spectrometry
- PDX:
-
Patient-derived lung tumor xenografts
- IF:
-
Immunofluorescence
- SPR:
-
Surface plasmon resonance
- PLA:
-
Proximity ligation assay
- TKI:
-
Tyrosine kinase inhibitor
- MAPK:
-
Mitogen-activated protein kinase
- MKK3/6:
-
Mitogen-activated protein kinase-kinase 3/6
- YAP:
-
Yes associated protein 1
- STAT3:
-
Signal transducer and activator of transcription 3
- EGFR:
-
Epidermal growth factor receptor
- MTA:
-
Microtubule-targeting agent
- EMT:
-
Epithelial-mesenchymal transition
- PPI:
-
Protein–protein interaction
- CDK1:
-
Cyclin dependent kinase 1
- DNA:
-
Deoxyribonucleic acid
- RNA:
-
Ribonucleic acid
- qRT-PCR:
-
Quantitative real-time PCR
- ANOVA:
-
One-way analysis of variance
- TCGA:
-
The Cancer Genome Atlas
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F (2021) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 71:209–249. https://doi.org/10.3322/caac.21660
Herbst RS, Morgensztern D, Boshoff C (2018) The biology and management of non-small cell lung cancer. Nature 553:446–454. https://doi.org/10.1038/nature25183
Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA (2008) Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc 83:584–594. https://doi.org/10.4065/83.5.584
Filaire E, Dupuis C, Galvaing G, Aubreton S, Laurent H, Richard R, Filaire M (2013) Lung cancer: what are the links with oxidative stress, physical activity and nutrition. Lung Cancer 82:383–389. https://doi.org/10.1016/j.lungcan.2013.09.009
Klaunig JE (2018) Oxidative stress and cancer. Curr Pharm Des 24:4771–4778. https://doi.org/10.2174/1381612825666190215121712
Hu J, Yao H, Gan F, Tokarski A, Wang Y (2012) Interaction of OKL38 and p53 in regulating mitochondrial structure and function. PLoS ONE 7:e43362. https://doi.org/10.1371/journal.pone.0043362
Ong CK, Leong C, Tan PH, Van T, Huynh H (2007) The role of 5’ untranslated region in translational suppression of OKL38 mRNA in hepatocellular carcinoma. Oncogene 26:1155–1165. https://doi.org/10.1038/sj.onc.1209896
Riou P, Saffroy R, Comoy J, Gross-Goupil M, Thiéry JP, Emile JF, Azoulay D, Piatier-Tonneau D, Lemoine A, Debuire B (2002) Investigation in liver tissues and cell lines of the transcription of 13 genes mapping to the 16q24 region that are frequently deleted in hepatocellular carcinoma. Clin Cancer Res 8:3178–3186
Ong CK, Ng CY, Leong C, Ng CP, Foo KT, Tan PH, Huynh H (2004) Genomic structure of human OKL38 gene and its differential expression in kidney carcinogenesis. J Biol Chem 279:743–754. https://doi.org/10.1074/jbc.M308668200
Liu M, Li Y, Chen L, Chan TH, Song Y, Fu L, Zeng TT, Dai YD, Zhu YH, Li Y et al (2014) Allele-specific imbalance of oxidative stress-induced growth inhibitor 1 associates with progression of hepatocellular carcinoma. Gastroenterology 146:1084–1096. https://doi.org/10.1053/j.gastro.2013.12.041
Wang G, Zhou H, Strulovici-Barel Y, Al-Hijji M, Ou X, Salit J, Walters MS, Staudt MR, Kaner RJ, Crystal RG (2017) Role of OSGIN1 in mediating smoking-induced autophagy in the human airway epithelium. Autophagy 13:1205–1220. https://doi.org/10.1080/15548627.2017.1301327
Khoi CS, Xiao CQ, Hung KY, Lin TY, Chiang CK (2022) Oxidative stress-induced growth inhibitor (OSGIN1), a target of X-box-binding protein 1, protects palmitic acid-induced Vascular lipotoxicity through maintaining autophagy. Biomedicines. https://doi.org/10.3390/biomedicines10050992
Mariani M, Karki R, Spennato M, Pandya D, He S, Andreoli M, Fiedler P, Ferlini C (2015) Class III β-tubulin in normal and cancer tissues. Gene 563:109–114. https://doi.org/10.1016/j.gene.2015.03.061
Borys F, Tobiasz P, Poterała M, Krawczyk H (2021) Development of novel derivatives of stilbene and macrocyclic compounds as potent of anti-microtubule factors. Biomed Pharmacother 133:110973. https://doi.org/10.1016/j.biopha.2020.110973
Tangutur AD, Kumar D, Krishna KV, Kantevari S (2017) Microtubule targeting agents as cancer chemotherapeutics: an overview of molecular hybrids as stabilizing and destabilizing agents. Curr Top Med Chem 17:2523–2537. https://doi.org/10.2174/1568026617666170104145640
Hinsch A, Chaker A, Burdelski C, Koop C, Tsourlakis MC, Steurer S, Rink M, Eichenauer TS, Wilczak W, Wittmer C et al (2017) βIII-tubulin overexpression is linked to aggressive tumor features and genetic instability in urinary bladder cancer. Hum Pathol 61:210–220. https://doi.org/10.1016/j.humpath.2016.11.005
Ti SC, Pamula MC, Howes SC, Duellberg C, Cade NI, Kleiner RE, Forth S, Surrey T, Nogales E, Kapoor TM (2016) Mutations in human tubulin proximal to the kinesin-binding site alter dynamic instability at microtubule plus- and minus-ends. Dev Cell 37:72–84. https://doi.org/10.1016/j.devcel.2016.03.003
Kanakkanthara A, Miller JH (2021) βIII-tubulin overexpression in cancer: causes, consequences, and potential therapies. Biochim Biophys Acta Rev Cancer 1876:188607. https://doi.org/10.1016/j.bbcan.2021.188607
McCarroll JA, Gan PP, Erlich RB, Liu M, Dwarte T, Sagnella SS, Akerfeldt MC, Yang L, Parker AL, Chang MH et al (2015) TUBB3/βIII-tubulin acts through the PTEN/AKT signaling axis to promote tumorigenesis and anoikis resistance in non-small cell lung cancer. Cancer Res 75:415–425. https://doi.org/10.1158/0008-5472.Can-14-2740
Huang J, Lan X, Wang T, Lu H, Cao M, Yan S, Cui Y, Jia D, Cai L, Xing Y (2020) Targeting the IL-1β/EHD1/TUBB3 axis overcomes resistance to EGFR-TKI in NSCLC. Oncogene 39:1739–1755. https://doi.org/10.1038/s41388-019-1099-5
Fourest-Lieuvin A, Peris L, Gache V, Garcia-Saez I, Juillan-Binard C, Lantez V, Job D (2006) Microtubule regulation in mitosis: tubulin phosphorylation by the cyclin-dependent kinase Cdk1. Mol Biol Cell 17:1041–1050. https://doi.org/10.1091/mbc.e05-07-0621
Janke C, Magiera MM (2020) The tubulin code and its role in controlling microtubule properties and functions. Nat Rev Mol Cell Biol 21:307–326. https://doi.org/10.1038/s41580-020-0214-3
Breitkopf SB, Asara JM (2012) Determining in vivo phosphorylation sites using mass spectrometry. Curr Protoc Mol Biol Chapter 18:Unit18.19.1–27. https://doi.org/10.1002/0471142727.mb1819s98
Yeung YT, Yin S, Lu B, Fan S, Yang R, Bai R, Zhang C, Bode AM, Liu K, Dong Z (2018) Losmapimod overcomes gefitinib resistance in non-small cell lung cancer by preventing tetraploidization. EBioMedicine 28:51–61. https://doi.org/10.1016/j.ebiom.2018.01.017
Chandrashekar DS, Karthikeyan SK, Korla PK, Patel H, Shovon AR, Athar M, Netto GJ, Qin ZS, Kumar S, Manne U et al (2022) UALCAN: an update to the integrated cancer data analysis platform. Neoplasia 25:18–27. https://doi.org/10.1016/j.neo.2022.01.001
Győrffy B, Surowiak P, Budczies J, Lánczky A (2013) Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS ONE 8:e82241. https://doi.org/10.1371/journal.pone.0082241
Kanehisa M, Sato Y, Kawashima M (2022) KEGG mapping tools for uncovering hidden features in biological data. Protein Sci 31:47–53. https://doi.org/10.1002/pro.4172
Lánczky A, Győrffy B (2021) Web-based survival analysis tool tailored for medical research (KMplot): development and implementation. J Med Internet Res 23:e27633. https://doi.org/10.2196/27633
Yan Y, Tao H, He J, Huang SY (2020) The HDOCK server for integrated protein-protein docking. Nat Protoc 15:1829–1852. https://doi.org/10.1038/s41596-020-0312-x
Huynh H, Ng CY, Ong CK, Lim KB, Chan TW (2001) Cloning and characterization of a novel pregnancy-induced growth inhibitor in mammary gland. Endocrinology 142:3607–3615. https://doi.org/10.1210/endo.142.8.8297
Duly AMP, Kao FCL, Teo WS, Kavallaris M (2022) βIII-tubulin gene regulation in health and disease. Front Cell Dev Biol 10:851542. https://doi.org/10.3389/fcell.2022.851542
Alfano A, Xu J, Yang X, Deshmukh D, Qiu Y (2022) SRC kinase-mediated tyrosine phosphorylation of TUBB3 regulates its stability and mitotic spindle dynamics in prostate cancer cells. Pharmaceutics. https://doi.org/10.3390/pharmaceutics14050932
Vona R, Mileo AM, Matarrese P (2021) Microtubule-based mitochondrial dynamics as a valuable therapeutic target in cancer. Cancers (Basel). https://doi.org/10.3390/cancers13225812
Zhang W, Cho WC, Bloukh SH, Edis Z, Du W, He Y, Hu HY, Hagen T, Falahati M (2022) An overview on the exploring the interaction of inorganic nanoparticles with microtubules for the advancement of cancer therapeutics. Int J Biol Macromol 212:358–369. https://doi.org/10.1016/j.ijbiomac.2022.05.150
Chin TM, Boopathy GTK, Man EPS, Clohessy JG, Csizmadia E, Quinlan MP, Putti T, Wan SC, Xie C, Ali A et al (2020) Targeting microtubules sensitizes drug resistant lung cancer cells to lysosomal pathway inhibitors. Theranostics 10:2727–2743. https://doi.org/10.7150/thno.38729
Gonçalves A, Braguer D, Kamath K, Martello L, Briand C, Horwitz S, Wilson L, Jordan MA (2001) Resistance to Taxol in lung cancer cells associated with increased microtubule dynamics. Proc Natl Acad Sci USA 98:11737–11742. https://doi.org/10.1073/pnas.191388598
Dhiman A, Sharma R, Singh RK (2022) Target-based anticancer indole derivatives and insight into structure-activity relationship: a mechanistic review update (2018–2021). Acta Pharm Sin B 12:3006–3027. https://doi.org/10.1016/j.apsb.2022.03.021
Cheng SS, Yang GJ, Wang W, Leung CH, Ma DL (2020) The design and development of covalent protein-protein interaction inhibitors for cancer treatment. J Hematol Oncol 13:26. https://doi.org/10.1186/s13045-020-00850-0
Ori-McKenney KM, McKenney RJ, Huang HH, Li T, Meltzer S, Jan LY, Vale RD, Wiita AP, Jan YN (2016) Phosphorylation of β-tubulin by the Down syndrome kinase, minibrain/DYRK1a, regulates microtubule dynamics and dendrite morphogenesis. Neuron 90:551–563. https://doi.org/10.1016/j.neuron.2016.03.027
Liu T, Wang Y, Wang J, Ren C, Chen H, Zhang J (2022) DYRK1A inhibitors for disease therapy: current status and perspectives. Eur J Med Chem 229:114062. https://doi.org/10.1016/j.ejmech.2021.114062
de Souza MM, Cenci AR, Teixeira KF, Machado V, Mendes Schuler MCG, Gonçalves AE, Paula Dalmagro A, André Cazarin C, Gomes Ferreira LL, de Oliveira AS, Andricopulo AD (2023) DYRK1A inhibitors and perspectives for the treatment of Alzheimer’s disease. Curr Med Chem 30:669–688. https://doi.org/10.2174/0929867329666220620162018
Lindberg MF, Meijer L (2021) Dual-specificity, tyrosine phosphorylation-regulated kinases (DYRKs) and cdc2-like kinases (CLKs) in human disease, an overview. Int J Mol Sci. https://doi.org/10.3390/ijms22116047
Reita D, Pabst L, Pencreach E, Guérin E, Dano L, Rimelen V, Voegeli AC, Vallat L, Mascaux C, Beau-Faller M (2021) Molecular mechanism of EGFR-TKI resistance in EGFR-mutated non-small cell lung cancer: application to biological diagnostic and monitoring. Cancers (Basel). https://doi.org/10.3390/cancers13194926
Čermák V, Dostál V, Jelínek M, Libusová L, Kovář J, Rösel D, Brábek J (2020) Microtubule-targeting agents and their impact on cancer treatment. Eur J Cell Biol 99:151075. https://doi.org/10.1016/j.ejcb.2020.151075
Ko JC, Chiu HC, Wo TY, Huang YJ, Tseng SC, Huang YC, Chen HJ, Syu JJ, Chen CY, Jian YT et al (2013) Inhibition of p38 MAPK-dependent MutS homologue-2 (MSH2) expression by metformin enhances gefitinib-induced cytotoxicity in human squamous lung cancer cells. Lung Cancer 82:397–406. https://doi.org/10.1016/j.lungcan.2013.09.011
Tung CL, Chiu HC, Jian YJ, Jian YT, Chen CY, Syu JJ, Wo TY, Huang YJ, Tseng SC, Lin YW (2014) Down-regulation of MSH2 expression by an Hsp90 inhibitor enhances pemetrexed-induced cytotoxicity in human non-small-cell lung cancer cells. Exp Cell Res 322:345–354. https://doi.org/10.1016/j.yexcr.2014.02.002
Greenberg AK, Basu S, Hu J, Yie TA, Tchou-Wong KM, Rom WN, Lee TC (2002) Selective p38 activation in human non-small cell lung cancer. Am J Respir Cell Mol Biol 26:558–564. https://doi.org/10.1165/ajrcmb.26.5.4689
Campbell RM, Anderson BD, Brooks NA, Brooks HB, Chan EM, De Dios A, Gilmour R, Graff JR, Jambrina E, Mader M et al (2014) Characterization of LY2228820 dimesylate, a potent and selective inhibitor of p38 MAPK with antitumor activity. Mol Cancer Ther 13:364–374. https://doi.org/10.1158/1535-7163.Mct-13-0513
Acknowledgements
We thank Xiangyu Wang for providing several plasmids. We are grateful to Dr. Xueli Tian for assisting with the SPR analysis and Ran Yang for preparation of immunohistochemical sections.
Funding
This work was supported by National Natural Science Foundation of Henan, China [Grant number 82103193, 82073075 and 212300410315], Major Science and Technology Projects in Henan Province [Grant number 221100310100], and the Bio & Medical Technology Development Program of the National Research Foundation of Korea (NRF) funded by the Ministry of Science & ICT [Grant number RS-2023-00237259].
Author information
Authors and Affiliations
Contributions
XX prepared the manuscript and performed the in vitro, the cell based and in vivo experiments; WN performed mass spectrometry, and phosphorproteomics analysis; JL performed computer modeling; KVL, YWY and KL performed data analysis and manuscript editing; YSS, ZD and DJK supervised the overall experimental design and provided the idea.
Corresponding authors
Ethics declarations
Conflict of interest
None of the authors have any competing interests.
Ethics approval and consent to participate
Written informed consent was obtained from all subjects. All animal experiments were conducted in agreement with the Guide for the Care and Use of Laboratory Animals and approved by the Ethics Committee of Zhengzhou University (Zhengzhou, Henan, China).
Consent for publication
All authors agreed on the manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.







Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xie, X., Laster, K.V., Li, J. et al. OSGIN1 is a novel TUBB3 regulator that promotes tumor progression and gefitinib resistance in non-small cell lung cancer. Cell. Mol. Life Sci. 80, 272 (2023). https://doi.org/10.1007/s00018-023-04931-4
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-023-04931-4