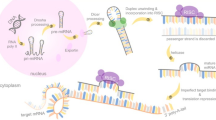
Abstract
Background
Increasing evidences has indicated that primary and acquired resistance of ovarian cancer (OC) to platinum is mediated by multiple molecular and cellular factors. Understanding these mechanisms could promote the therapeutic efficiency for patients with OC.
Methods
Here, we screened the expression pattern of circRNAs in samples derived from platinum-resistant and platinum-sensitive OC patients using RNA-sequencing (RNA-seq). The expression of hsa_circ_0010467 was validated by Sanger sequencing, RT-qPCR, and fluorescence in situ hybridization (FISH) assays. Overexpression and knockdown experiments were performed to explore the function of hsa_circ_0010467. The effects of hsa_circ_0010467 on enhancing platinum treatment were validated in OC cells, mouse model and patient-derived organoid (PDO). RNA pull-down, RNA immunoprecipitation (RIP), and dual-luciferase reporter assays were performed to investigate the interaction between hsa_circ_0010467 and proteins.
Results
Increased expression of hsa_circ_0010467 is observed in platinum-resistant OC cells, tissues and serum exosomes, which is positively correlated with advanced tumor stage and poor prognosis of OC patients. Hsa_circ_0010467 is found to maintain the platinum resistance via inducing tumor cell stemness, and silencing hsa_circ_0010467 substantially increases the efficacy of platinum treatment on inhibiting OC cell proliferation. Further investigation reveals that hsa_circ_0010467 acts as a miR-637 sponge to mediate the repressive effect of miR-637 on leukemia inhibitory factor (LIF) and activates the LIF/STAT3 signaling pathway. We further discover that AUF1 could promote the biogenesis of hsa_circ_0010467 in OC.
Conclusion
Our study uncovers the mechanism that hsa_circ_0010467 mediates the platinum resistance of OC through AUF1/hsa_circ_0010467/miR-637/LIF/STAT3 axis, and provides potential targets for the treatment of platinum-resistant OC patients.








Similar content being viewed by others
Availability of data and material
Raw RNA-seq data are deposited in the Gene Expression Omnibus (GEO) database under the accession number of GSE214302. Software and resources used for data analysis and visualization are described in the method section.
References
Momenimovahed Z, Tiznobaik A, Taheri S, Salehiniya H (2019) Ovarian cancer in the world: epidemiology and risk factors. Int J Womens Health 11:287–299
Budiana ING, Angelina M, Pemayun TGA (2019) Ovarian cancer: Pathogenesis and current recommendations for prophylactic surgery. J Turk Ger Gynecol Assoc 20(1):47–54
Matulonis UA, Sood AK, Fallowfield L, Howitt BE, Sehouli J, Karlan BY (2016) Ovarian cancer. Nat Rev Dis Primers 2:16061
Jeck WR, Sharpless NE (2014) Detecting and characterizing circular RNAs. Nat Biotechnol 32(5):453–461
Li S, Han L (2019) Circular RNAs as promising biomarkers in cancer: detection, function, and beyond. Genome Med 11(1):15
Fang Z, Jiang C, Li S (2020) The potential regulatory roles of circular RNAs in tumor immunology and immunotherapy. Front Immunol 11:617583
Ruan H, Xiang Y, Ko J, Li S, Jing Y, Zhu X, Ye Y, Zhang Z, Mills T, Feng J et al (2019) Comprehensive characterization of circular RNAs in ~ 1000 human cancer cell lines. Genome Med 11(1):55
Liang WC, Wong CW, Liang PP, Shi M, Cao Y, Rao ST, Tsui SK, Waye MM, Zhang Q, Fu WM et al (2019) Translation of the circular RNA circbeta-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol 20(1):84
Sheng R, Li X, Wang Z, Wang X (2020) Circular RNAs and their emerging roles as diagnostic and prognostic biomarkers in ovarian cancer. Cancer Lett 473:139–147
Gan X, Zhu H, Jiang X, Obiegbusi SC, Yong M, Long X, Hu J (2020) CircMUC16 promotes autophagy of epithelial ovarian cancer via interaction with ATG13 and miR-199a. Mol Cancer 19(1):45
Zhang L, Zhou Q, Qiu Q, Hou L, Wu M, Li J, Li X, Lu B, Cheng X, Liu P et al (2019) CircPLEKHM3 acts as a tumor suppressor through regulation of the miR-9/BRCA1/DNAJB6/KLF4/AKT1 axis in ovarian cancer. Mol Cancer 18(1):144
Zhao Z, Ji M, Wang Q, He N, Li Y (2019) Circular RNA Cdr1as upregulates SCAI to suppress cisplatin resistance in ovarian cancer via miR-1270 suppression. Mol Ther Nucleic Acids 18:24–33
Li Z, Zhang J, Liu X, Li S, Wang Q, Di C, Hu Z, Yu T, Ding J, Li J et al (2018) The LINC01138 drives malignancies via activating arginine methyltransferase 5 in hepatocellular carcinoma. Nat Commun 9(1):1572
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120
Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12(4):357–360
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R (2009) Genome project data processing S: The sequence alignment/Map format and SAMtools. Bioinformatics 25(16):2078–2079
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M et al (2013) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495(7441):333–338
Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, Celniker SE, Graveley BR, Lai EC (2014) Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep 9(5):1966–1980
Gao Y, Zhang J, Zhao F (2018) Circular RNA identification based on multiple seed matching. Brief Bioinform 19(5):803–810
Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L (2014) Complementary sequence-mediated exon circularization. Cell 159(1):134–147
Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26(6):841–842
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15(12):550
Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL (2015) StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33(3):290–295
Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, Feng T, Zhou L, Tang W, Zhan L et al (2021) clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation (Camb) 2(3):100141
Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P (2015) The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst 1(6):417–425
Kopper O, de Witte CJ, Lohmussaar K, Valle-Inclan JE, Hami N, Kester L, Balgobind AV, Korving J, Proost N, Begthel H et al (2019) An organoid platform for ovarian cancer captures intra- and interpatient heterogeneity. Nat Med 25(5):838–849
Glazar P, Papavasileiou P, Rajewsky N (2014) circBase: a database for circular RNAs. RNA 20(11):1666–1670
Nabet BY, Qiu Y, Shabason JE, Wu TJ, Yoon T, Kim BC, Benci JL, DeMichele AM, Tchou J, Marcotrigiano J et al (2017) Exosome RNA unshielding couples stromal activation to pattern recognition receptor signaling in cancer. Cell 170(2):352-366e313
Kamerkar S, LeBleu VS, Sugimoto H, Yang S, Ruivo CF, Melo SA, Lee JJ, Kalluri R (2017) Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature 546(7659):498–503
Kahlert C, Kalluri R (2013) Exosomes in tumor microenvironment influence cancer progression and metastasis. J Mol Med (Berl) 91(4):431–437
Shafee N, Smith CR, Wei S, Kim Y, Mills GB, Hortobagyi GN, Stanbridge EJ, Lee EY (2008) Cancer stem cells contribute to cisplatin resistance in Brca1/p53-mediated mouse mammary tumors. Cancer Res 68(9):3243–3250
Zhang Y, Wang Z, Yu J, Shi J, Wang C, Fu W, Chen Z, Yang J (2012) Cancer stem-like cells contribute to cisplatin resistance and progression in bladder cancer. Cancer Lett 322(1):70–77
Wen Y, Hou Y, Yi X, Sun S, Guo J, He X, Li T, Cai J, Wang Z (2021) EZH2 activates CHK1 signaling to promote ovarian cancer chemoresistance by maintaining the properties of cancer stem cells. Theranostics 11(4):1795–1813
Yokoyama Y, Zhu H, Lee JH, Kossenkov AV, Wu SY, Wickramasinghe JM, Yin X, Palozola KC, Gardini A, Showe LC et al (2016) BET inhibitors suppress ALDH activity by targeting ALDH1A1 super-enhancer in ovarian cancer. Cancer Res 76(21):6320–6330
Ramadoss S, Sen S, Ramachandran I, Roy S, Chaudhuri G, Farias-Eisner R (2017) Lysine-specific demethylase KDM3A regulates ovarian cancer stemness and chemoresistance. Oncogene 36(11):1537–1545
Laury AR, Hornick JL, Perets R, Krane JF, Corson J, Drapkin R, Hirsch MS (2010) PAX8 reliably distinguishes ovarian serous tumors from malignant mesothelioma. Am J Surg Pathol 34(5):627–635
Jia L, Wang Y, Wang CY (2021) circFAT1 Promotes cancer stemness and immune evasion by promoting STAT3 activation. Adv Sci (Weinh) 8(13):2003376
Chen MW, Yang ST, Chien MH, Hua KT, Wu CJ, Hsiao SM, Lin H, Hsiao M, Su JL, Wei LH (2017) The STAT3-miRNA-92-Wnt signaling pathway regulates spheroid formation and malignant progression in ovarian cancer. Cancer Res 77(8):1955–1967
Bareiss PM, Paczulla A, Wang H, Schairer R, Wiehr S, Kohlhofer U, Rothfuss OC, Fischer A, Perner S, Staebler A et al (2013) SOX2 expression associates with stem cell state in human ovarian carcinoma. Cancer Res 73(17):5544–5555
Condello S, Morgan CA, Nagdas S, Cao L, Turek J, Hurley TD, Matei D (2015) β-Catenin-regulated ALDH1A1 is a target in ovarian cancer spheroids. Oncogene 34(18):2297–2308
Liu M, Wang Q, Shen J, Yang BB, Ding X (2019) Circbank: a comprehensive database for circRNA with standard nomenclature. RNA Biol 16(7):899–905
Wu W, Ji P, Zhao F (2020) CircAtlas: an integrated resource of one million highly accurate circular RNAs from 1070 vertebrate transcriptomes. Genome Biol 21(1):101
Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M (2016) CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol 13(1):34–42
Hu C, Xia R, Zhang X, Li T, Ye Y, Li G, He R, Li Z, Lin Q, Zheng S et al (2022) circFARP1 enables cancer-associated fibroblasts to promote gemcitabine resistance in pancreatic cancer via the LIF/STAT3 axis. Mol Cancer 21(1):24
Zhang JF, He ML, Fu WM, Wang H, Chen LZ, Zhu X, Chen Y, Xie D, Lai P, Chen G et al (2011) Primate-specific microRNA-637 inhibits tumorigenesis in hepatocellular carcinoma by disrupting signal transducer and activator of transcription 3 signaling. Hepatology 54(6):2137–2148
Penuelas S, Anido J, Prieto-Sanchez RM, Folch G, Barba I, Cuartas I, Garcia-Dorado D, Poca MA, Sahuquillo J, Baselga J et al (2009) TGF-beta increases glioma-initiating cell self-renewal through the induction of LIF in human glioblastoma. Cancer Cell 15(4):315–327
Zheng X, Huang M, Xing L, Yang R, Wang X, Jiang R, Zhang L, Chen J (2020) The circRNA circSEPT9 mediated by E2F1 and EIF4A3 facilitates the carcinogenesis and development of triple-negative breast cancer. Mol Cancer 19(1):73
Tang Z, Kang B, Li C, Chen T, Zhang Z (2019) GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res 47(W1):W556–W560
Lyu X, Zeng L, Shi J, Ming Z, Li W, Liu B, Chen Y, Yuan B, Sun R, Yuan J et al (2022) Essential role for STAT3/FOXM1/ATG7 signaling-dependent autophagy in resistance to Icotinib. J Exp Clin Cancer Res 41(1):200
Guo R, Jiang M, Wang G, Li B, Jia X, Ai Y, Chen S, Tang P, Liu A, Yuan Q et al (2022) IL6 supports long-term expansion of hepatocytes in vitro. Nat Commun 13(1):7345
Chiba T, Yamada M, Hashimoto Y, Sato M, Sasabe J, Kita Y, Terashita K, Aiso S, Nishimoto I, Matsuoka M (2005) Development of a femtomolar-acting humanin derivative named colivelin by attaching activity-dependent neurotrophic factor to its N terminus: characterization of colivelin-mediated neuroprotection against Alzheimer’s disease-relevant insults in vitro and in vivo. J Neurosci 25(44):10252–10261
Zhou S, Dai Q, Huang X, Jin A, Yang Y, Gong X, Xu H, Gao X, Jiang L (2021) STAT3 is critical for skeletal development and bone homeostasis by regulating osteogenesis. Nat Commun 12(1):6891
Chen L, Kong R, Wu C, Wang S, Liu Z, Liu S, Li S, Chen T, Mao C, Liu S (2020) Circ-MALAT1 functions as both an mRNA translation brake and a microRNA sponge to promote self-renewal of hepatocellular cancer stem cells. Adv Sci (Weinh) 7(4):1900949
Gao W, Guo H, Niu M, Zheng X, Zhang Y, Xue X, Bo Y, Guan X, Li Z, Guo Y et al (2020) circPARD3 drives malignant progression and chemoresistance of laryngeal squamous cell carcinoma by inhibiting autophagy through the PRKCI-Akt-mTOR pathway. Mol Cancer 19(1):166
Feng ZH, Zheng L, Yao T, Tao SY, Wei XA, Zheng ZY, Zheng BJ, Zhang XY, Huang B, Liu JH et al (2021) EIF4A3-induced circular RNA PRKAR1B promotes osteosarcoma progression by miR-361-3p-mediated induction of FZD4 expression. Cell Death Dis 12(11):1025
Friedman RC, Farh KK, Burge CB, Bartel DP (2009) Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19(1):92–105
Que T, Song Y, Liu Z, Zheng S, Long H, Li Z, Liu Y, Wang G, Liu Y, Zhou J et al (2015) Decreased miRNA-637 is an unfavorable prognosis marker and promotes glioma cell growth, migration and invasion via direct targeting Akt1. Oncogene 34(38):4952–4963
Wang L, Jiang F, Xia X, Zhang B (2019) LncRNA FAL1 promotes carcinogenesis by regulation of miR-637/NUPR1 pathway in colorectal cancer. Int J Biochem Cell Biol 106:46–56
Spitzner M, Roesler B, Bielfeld C, Emons G, Gaedcke J, Wolff HA, Rave-Frank M, Kramer F, Beissbarth T, Kitz J et al (2014) STAT3 inhibition sensitizes colorectal cancer to chemoradiotherapy in vitro and in vivo. Int J Cancer 134(4):997–1007
Funding
This study was supported by the National Natural Science Foundation of China (81972431 and 32100517) and General Project of Natural Science Foundation of Shanghai (21ZR1415000).
Author information
Authors and Affiliations
Contributions
X.W. and S.L. conceived and supervised the project; Y.W., and Z.F. collected clinical samples; Y.W. and M.X. conducted the experiments; S.L. and W.H. performed the computational analysis; J.W., X. H., Y.W., S.C., F.X., and H.W. interpreted the results; Y.W., S.L., and X.W. wrote the manuscript with comments from all the other authors; All authors read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no potential conflicts of interest.
Ethics approval and consent to participate
All animal experiments were approved by the Animal Care Committee of Fudan University, and were performed in accordance with the light of NIH Guidelines for the Care and Use of Laboratory Animals. The Ethics Committee of Fudan University Shanghai Cancer Center approved the study, and all participants signed informed consent statements.
Consent for publication
All authors give consent for the publication of this manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wu, Y., Xu, M., Feng, Z. et al. AUF1-induced circular RNA hsa_circ_0010467 promotes platinum resistance of ovarian cancer through miR-637/LIF/STAT3 axis. Cell. Mol. Life Sci. 80, 256 (2023). https://doi.org/10.1007/s00018-023-04906-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-023-04906-5