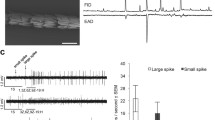
Abstract
Male moths utilize their pheromone communication systems to distinguish potential mates from other sympatric species, which contributes to maintaining reproductive isolation and even drives speciation. The molecular mechanisms underlying the evolution of pheromone communication systems are usually studied between closely-related moth species for their similar but divergent traits associated with pheromone production, detection, and/or processing. In this study, we first identified the functional differentiation in two orthologous pheromone receptors, OR14b, and OR16, in four Helicoverpa species, Helicoverpa armigera, H. assulta, H. zea, and H. gelotopoeon. To understand the substrate response specificity of these two PRs, we performed all-atom molecular dynamics simulations of OR14b and OR16 based on AlphaFold2 structural prediction, and molecular docking, allowing us to predict a few key amino acids involved in substrate binding. These candidate residues were further tested and validated by site-directed mutagenesis and functional analysis. These results together identified two hydrophobic amino acids at positions 164 and 232 are the determinants of the response specificity of HarmOR14b and HzeaOR14b to Z9-14:Ald and Z9-16:Ald by directly interacting with the substrates. Interestingly, in OR16 orthologs, we found that position 66 alone determines the specific binding of Z11-16:OH, likely via allosteric interactions. Overall, we have developed an effective integrated method to identify the critical residues for substrate selectivity of ORs and elucidated the molecular mechanism of the diversification of pheromone recognition systems.







Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in this article (and its Supplementary files).
Abbreviations
- OR:
-
Odorant receptor
- Orco:
-
Odorant receptor co-receptor
- PBP:
-
Pheromone binding protein
- SNMP:
-
Sensory neuron membrane protein
- PDE:
-
Pheromone degrading enzyme
- PR:
-
Pheromone receptor
- cRNA:
-
Caped RNA
- TMD:
-
Transmembrane domain
- ECL:
-
Extracellular loop
- cryoEM:
-
Cryogenic electron microscopy
- CGenFF:
-
CHARMM general force field
References
Cardé RT, Baker TC (1984) Sexual communication with pheromones. In: Bell WJ, Cardé RT (eds) Chemical ecology of insects. Springer, Boston, pp 355–383
Baker TC (2002) Mechanism for saltational shifts in pheromone communication systems. Proc Natl Acad Sci USA 99:13368–13370
Groot AT, Horovitz JL, Hamilton J, Santangelo RG, Schal C, Gould F (2006) Experimental evidence for interspecific directional selection on moth pheromone communication. Proc Natl Acad Sci USA 103:5858–5863
Smadja CM, Butlin RK (2009) On the scent of speciation: the chemosensory system and its role in premating isolation. Heredity 102:77–97
Löfstedt C (1993) Moth pheromone genetics and evolution. Philos Trans R Soc Lond B Biol Sci 340:167–177
Roelofs WL, Liu W, Hao G, Jiao H, Rooney AP Jr, Linn CE (2002) Evolution of moth sex pheromones via ancestral genes. Proc Natl Acad Sci USA 99:13621–13626
Hillier NK, Baker TC (2016) Pheromones of Heliothine moths. In: Allison JD, Cardé RT (eds) Pheromone communication in moths: evolution, behavior, and application. University of California Press, Berkeley, pp 301–333
Phelan PL (1992) Evolution of sex pheromones and the role of asymmetric tracking. In: Roitberg BD, Isman MB (eds) Insect chemical ecology: an evolutionary approach. Chapman and Hall, New York, pp 265–314
Phelan PL (1997) Genetic and phylogenetics in the evolution of sex pheromones. In: Cardé TR, Minks KA (eds) Insect pheromone research. Chapman and Hall, New York, pp 563–579
Jurenka RA, Subchev M, Abad J-L, Choi M-Y, Fabriàs G (2003) Sex pheromone biosynthetic pathway for disparlure in the gypsy moth, Lymantria dispar. Proc Natl Acad Sci USA 100:809–814
Yew JY, Chung H (2015) Insect pheromones: an overview of function, form, and discovery. Prog Lipid Res 59:88–105
Löfstedt C, Herrebout WM, Menken SBJ (1991) Sex pheromones and their potential role in the evolution of reproductive isolation in small ermine moths (Yponomeutidae). Chemoecology 2:20–28
Groot AT, Dekker T, Heckel DG (2016) The genetic basis of pheromone evolution in moths. Annu Rev Entomol 61:99–117
Lassance J-M, Groot AT, Liénard MA, Antony B, Borgwardt C, Andersson F, Hedenström E, Heckel DG, Löfstedt C (2010) Allelic variation in a fatty-acyl reductase gene causes divergence in moth sex pheromones. Nature 466:486–489
Lassance J-M, Liénard MA, Antony B, Qian S, Fujii T, Tabata J, Ishikawa Y, Löfstedt C (2013) Functional consequences of sequence variation in the pheromone biosynthetic gene pgFAR for Ostrinia moths. Proc Natl Acad Sci USA 110:3967–3972
Buček A, Matoušková P, Vogel H, Šebesta P, Jahn U, Weiβflog J, Svatoš A, Pichová I (2015) Evolution of moth sex pheromone composition by a single amino acid substitution in a fatty acid desaturase. Proc Natl Acad Sci USA 112:12586–12591
Grosse-Wilde E, Svatoš A, Krieger J (2006) A pheromone-binding protein mediates the bombykol-induced activation of a pheromone receptor in vitro. Chem Senses 31:547–555
Zhu G, Xu J, Cui Z, Dong X, Ye Z, Niu D, Huang Y, Dong S (2016) Functional characterization of SlitPBP3 in Spodoptera litura by CRISPR/Cas9 mediated genome editing. Insect Biochem Mol 75:1–9
Pregitzer P, Greschista M, Breer H, Krieger J (2014) The sensory neurone membrane protein SNMP1 contributes to the sensitivity of a pheromone detection system. Insect Mol Biol 23:733–742
Ishida Y, Leal WS (2005) Rapid inactivation of a moth pheromone. Proc Natl Acad Sci USA 102:14075–14079
Durand N, Carot-Sans G, Bozzolan F, Rosell G, Siaussat D, Debernard S, Chertemps T, Maïbèche-Coisne M (2011) Degradation of pheromone and plant volatile components by a same odorant-degrading enzyme in the cotton leafworm, Spodoptera littoralis. PLoS One 6:e29147
Sakurai T, Mitsuno H, Haupt SS, Uchino K, Yokohari F, Nishioka T, Kobayashi I, Sezutsu H, Tamura T, Kanzaki R (2011) A single sex pheromone receptor determines chemical response specificity of sexual behavior in the silkmoth Bombyx mori. PLoS Genet 7:e1002115
Chang H, Liu Y, Ai D, Jiang X, Dong S, Wang G (2017) A pheromone antagonist regulates optimal mating time in the moth Helicoverpa armigera. Curr Biol 27:1610–1615
Bastin-Héline L, de Fouchier A, Cao S, Koutroumpa F, Caballero-Vidal G, Robakiewicz S, Monsempes C, François M-C, Ribeyre T, Maria A et al (2019) A novel lineage of candidate pheromone receptors for sex communication in moths. Elife 8:e49826
Leary GP, Allen JE, Bunger PL, Luginbill JB Jr, Linn CE, Macallister IE, Kavanaugh MP, Wanner KW (2012) Single mutation to a sex pheromone receptor provides adaptive specificity between closely related moth species. Proc Natl Acad Sci USA 109:14081–14086
Yang K, Huang L, Ning C, Wang C (2017) Two single-point mutations shift the ligand selectivity of a pheromone receptor between two closely related moth species. Elife 6:e29100
Krieger J, Grosse-Wilde E, Gohl T, Dewer Y, Raming K, Breer H (2004) Genes encoding candidate pheromone receptors in a moth (Heliothis virescens). Proc Natl Acad Sci USA 101:11845–11850
Grosse-Wilde E, Kuebler LS, Bucks S, Vogel H, Wicher D, Hansson BS (2011) Antennal transcriptome of Manduca sexta. Proc Natl Acad Sci USA 108:7449–7454
Liu Y, Gu S, Zhang Y, Guo Y, Wang G (2012) Candidate olfaction genes identified within the Helicoverpa armigera antennal transcriptome. PLoS One 7:e48260
Yuvaraj JK, Andersson MN, Zhang D, Löfstedt C (2018) Antennal transcriptome analysis of the chemosensory gene families from trichoptera and basal Lepidoptera. Front Physiol 9:1365
Sakurai T, Nakagawa T, Mitsuno H, Mori H, Endo Y, Tanoue S, Yasukochi Y, Touhara K, Nishioka T (2004) Identification and functional characterization of a sex pheromone receptor in the silkmoth Bombyx mori. Proc Natl Acad Sci USA 101:16653–16658
Nakagawa T, Sakurai T, Nishioka T, Touhara K (2005) Insect sex-pheromone signals mediated by specific combinations of olfactory receptors. Science 307:1638–1642
Liu Y, Liu C, Lin K, Wang G (2013) Functional specificity of sex pheromone receptors in the cotton bollworm Helicoverpa armigera. PLoS One 8:e62094
Yuvaraj JK, Andersson MN, Corcoran JA, Anderbrant O, Löfstedt C (2018) Functional characterization of odorant receptors from Lampronia capitella suggests a non-ditrysian origin of the lepidopteran pheromone receptor clade. Insect Biochem Mol 100:39–47
Fitt GP (1989) The ecology of Heliothis species in relation to agroecosystems. Annu Rev Entomol 34:17–53
Behere G, Tay WT, Russell DA, Batterham P (2008) Molecular markers to discriminate among four pest species of Helicoverpa (Lepidoptera: Noctuidae). Bull Entomol Res 98:599–603
Wang C, Dong J (2001) Interspecific hybridization of Helicoverpa armigera and H. assulta (Lepidoptera: Noctuidae). Chin Sci Bull 46:489–491
Klun JA, Plimmer JR, Bierl-Leonhardt BA, Sparks AN, Primiani M, Chapman OL, Lee GH, Lepone G (1980) Sex pheromone chemistry of female corn earworm moth, Heliothis zea. J Chem Ecol 6:165–175
Pope MM, Gaston LK, Baker TC (1984) Composition, quantification, and periodicity of sex pheromone volatiles from individual Heliothis zea females. J Insect Physiol 30:943–945
Kehat M, Dunkelblum E (1990) Behavioral responses of male Heliothis armigera (Lepidoptera: Noctuidae) moths in a flight tunnel to combinations of components identified from female sex pheromone glands. J Insect Behav 3:75–83
Vickers NJ, Christensen TA, Mustaparta H, Baker TC (1991) Chemical communication in heliothine moths III. Flight behavior of male Helicoverpa zea and Heliothis virescens in ratios of intra- and interspecific sex pheromone components. J Comp Physiol A 169:275–280
Cork A, Boo KS, Dunkelblum E, Hall DR, Jee-Rajunga K, Kehat M, Jie EK, Park KC, Tepgidagarn P, Xun L (1992) Female sex pheromone of oriental tobacco budworm, Helicoverpa assulta (Guenee) (Lepidoptera: Noctuidae): identification and field testing. J Chem Ecol 18:403–418
Boo KS, Park KC, Hall DR, Cork A, Berg BG, Mustaparta H (1995) (Z)-9-tetradecenal: a potent inhibitor of pheromone-mediated communication in the oriental tobacco budworm moth, Helicoverpa assulta. J Comp Physiol A 177:695–699
Fadamiro HY, Baker TC (1997) Helicoverpa zea males (Lepidoptera: Noctuidae) respond to the intermittent fine structure of their sex pheromone plume and an antagonist in a flight tunnel. Physiol Entomol 22:316–324
Quero C, Baker TC (1999) Antagonistic effect of (Z)-11-Hexadecen-1-ol on the pheromone-mediated flight of Helicoverpa zea (Boddie) (Lepidoptera: Noctuidae). J Insect Behav 12:701–710
Cork A, Lobos EA (2003) Female sex pheromone components of Helicoverpa gelotopoeon: first heliothine pheromone without (Z)-11-hexadecenal. Entomol Exp Appl 107:201–206
Zhang J, Salcedo C, Fang Y, Zhang R, Zhang Z (2012) An overlooked component: (Z)-9-tetradecenal as a sex pheromone in Helicoverpa armigera. J Insect Physiol 58:1209–1216
Wu H, Xu M, Hou C, Huang L, Dong J, Wang C (2015) Specific olfactory neurons and glomeruli are associated to differences in behavioral responses to pheromone components between two Helicoverpa species. Front Behav Neurosci 9:206
Zhang J, Wang B, Dong S, Cao D, Dong J, Walker WB, Liu Y, Wang G (2015) Antennal transcriptome analysis and comparison of chemosensory gene families in two closely related noctuidae moths, Helicoverpa armigera and H. assulta. PLoS One 10:e0117054
Chang H, Guo M, Wang B, Liu Y, Dong S, Wang G (2016) Sensillar expression and responses of olfactory receptors reveal different peripheral coding in two Helicoverpa species using the same pheromone components. Sci Rep 6:18742
Zhu J, Ban L, Song L, Liu Y, Pelosi P, Wang G (2016) General odorant-binding proteins and sex pheromone guide larvae of Plutella xylostella to better food. Insect Biochem Mol 72:10–19
Gao M, Nakajima An D, Parks JM, Skolnick J (2022) AF2Complex predicts direct physical interactions in multimeric proteins with deep learning. Nat Commun 13:1744
Eberhardt J, Santos-Martins D, Tillack AF, Forli S (2021) AutoDock Vina 1.2.0: new docking methods, expanded force field, and python bindings. J Chem Inf Model 61:3891–3898
Jo S, Kim T, Iyer VG, Im W (2008) CHARMM-GUI: a web-based graphical user interface for CHARMM. J Comput Chem 29:1859–1865
Huang J, Rauscher S, Nawrocki G, Ran T, Feig M, de Groot BL, Grubmüller H Jr, MacKerell AD (2017) CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat Methods 14:71–73
Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, Darian E, Guvench O, Lopes P, Vorobyov I et al (2010) CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J Comput Chem 31:671–690
Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E (2015) GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2:19–25
Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graph 14:33–38
Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A et al (2021) Highly accurate protein structure prediction with AlphaFold. Nature 596:583–589
Yan H, Jafari S, Pask G, Zhou X, Reinberg D, Desplan C (2020) Evolution, developmental expression and function of odorant receptors in insects. J Exp Biol 223:jeb208215
del Mármol J, Yedlin MA, Ruta V (2021) The structural basis of odorant recognition in insect olfactory receptors. Nature 597:126–131
Cossé AA, Todd JL, Baker TC (1998) Neurons discovered in male Helicoverpa zea antennae that correlate with pheromone-mediated attraction and interspecific antagonism. J Comp Physiol A 182:585–594
Guo H, Huang L, Gong X, Wang C (2022) Comparison of functions of pheromone receptor repertoires in Helicoverpa armigera and Helicoverpa assulta using a Drosophila expression system. Insect Biochem Mol 141:103702
Butterwick JA, del Mármol J, Kim KH, Kahlson MA, Rogow JA, Walz T, Ruta V (2018) Cryo-EM structure of the insect olfactory receptor Orco. Nature 560:447–452
Stansfeld PJ, Sansom MS (2011) Molecular simulation approaches to membrane proteins. Structure 19:1562–1572
Wang Y, Bugge K, Kragelund BB, Lindorff-Larsen K (2018) Role of protein dynamics in transmembrane receptor signalling. Curr Opin Struct Biol 48:74–82
Pellegrino M, Steinbach N, Stensmyr MC, Hansson BS, Vosshall LB (2011) A natural polymorphism alters odour and DEET sensitivity in an insect odorant receptor. Nature 478:511–514
Hughes DT, Wang G, Zwiebel LJ, Luetje CW (2014) A determinant of odorant specificity is located at the extracellular loop 2-transmembrane domain 4 interface of an Anopheles gambiae odorant receptor subunit. Chem Senses 39:761–769
Cao S, Liu Y, Wang B, Wang G (2021) A single point mutation causes one-way alteration of pheromone receptor function in two Heliothis species. iScience 24:102981
Cho S, Mitchell A, Mitter C, Regier J, Matthews M, Robertson R (2008) Molecular phylogenetics of heliothine moths (Lepidoptera: Noctuidae: Heliothinae), with comments on the evolution of host range and pest status. Syst Entomol 33:581–594
Laster ML, Sheng CF (1995) Search for hybrid sterility for Helicoverpa zea in crosses between the north American H. zea and H. armigera (Lepidoptera: Noctuidae) from China. J Econ Entomol 88:1288–1291
Anderson CJ, Oakeshott JG, Tay WT, Gordon KHJ, Zwick A, Walsh TK (2018) Hybridization and gene flow in the mega-pest lineage of moth, Helicoverpa. Proc Natl Acad Sci USA 115:5034–5039
Pogue MG (2004) A new synonym of Helicoverpa zea (Boddie) and differentiation of adult males of H. zea and H. armigera (Hübner) (Lepidoptera: Noctuidae: Heliothinae). Ann Entomol Soc Am 97:1222–1226
Mallet J, Korman A, Heckel DG, King P (1993) Biochemical genetics of Heliothis and Helicoverpa (Lepidoptera: Noctuidae) and evidence for a founder event in Helicoverpa zea. Ann Entomol Soc Am 86:189–197
Pearce SL, Clarke DF, East PD, Elfekih S, Gordon KHJ, Jermiin LS, McGaughran A, Oakeshott JG, Papanikolaou A, Perera OP et al (2017) Genomic innovations, transcriptional plasticity and gene loss underlying the evolution and divergence of two highly polyphagous and invasive Helicoverpa pest species. BMC Biol 15:63
Acknowledgements
We thank Dr. Fred Gould (North Carolina State University, Raleigh, NC, USA) for providing the experimental materials and comments. We also thank Dr. Emmanuelle Jacquin-Joly (INRAE, Sorbonne Universite, CNRS, IRD, UPEC, Universite de Paris, Institute of Ecology and Environmental Sciences of Paris) for helpful discussions. Y.W acknowledges the access to computational resources from the Information Technology Center and State Key Lab of CAD&CG, Zhejiang University.
Funding
This work was funded by National Natural Science Foundation of China (32130089 to G.W., 32272540, 32072509 to Y.L., and 11932017 to P.X.), the National Key R&D Program of China (No. 2021YFF1200404 to Y.W.), the Fundamental Research Funds for the Central Universities of China (No. K20220228 to Y.W.), China Postdoctoral Science Foundation (2020M680785 to S.C.), and the Shenzhen Science and Technology Program (Grant No. KQTD20180411143628272 to G.W.), Central Public-interest Scientific Institution Basal Research Fund (CAAS-ZDRW202108), Projects subsidized by Special Funds for Science Technology Innovation and Industrial Development of Shenzhen Dapeng New District (PT202101-02), and the Agricultural Science and Technology Innovation Program (ASTIP). The funder had no role in study design, data collection and interpretation or manuscript preparation.
Author information
Authors and Affiliations
Contributions
Conceptualization, YL, GW and YW; conducted experiments, SC, YL, CS and BW; data analysis and visualization, SC and CS; writing—original draft, SC, YL and CS; writing—review and editing, SC, CS, BW, PX, YW, YL and GW; funding acquisition, SC, PX, YW, YL and GW.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Cao, S., Shi, C., Wang, B. et al. Evolutionary shifts in pheromone receptors contribute to speciation in four Helicoverpa species. Cell. Mol. Life Sci. 80, 199 (2023). https://doi.org/10.1007/s00018-023-04837-1
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-023-04837-1