Abstract
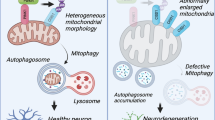
Ufmylation is a recently identified small ubiquitin-like modification, whose biological function and relevant cellular targets are poorly understood. Here we present evidence of a neuroprotective role for Ufmylation involving Autophagy-related gene 9 (Atg9) during Drosophila aging. The Ufm1 system ensures the health of aged neurons via Atg9 by coordinating autophagy and mTORC1, and maintaining mitochondrial homeostasis and JNK (c-Jun N-terminal kinase) activity. Neuron-specific expression of Atg9 suppresses the age-associated movement defect and lethality caused by loss of Ufmylation. Furthermore, Atg9 is identified as a conserved target of Ufm1 conjugation mediated by Ddrgk1, a critical regulator of Ufmylation. Mammalian Ddrgk1 was shown to be indispensable for the stability of endogenous Atg9A protein in mouse embryonic fibroblast (MEF) cells. Taken together, our findings might have important implications for neurodegenerative diseases in mammals.







Similar content being viewed by others
Availability of data and materials
No new datasets were generated during the current study.
References
Klionsky DJ et al (2016) Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12(1):1–222. https://doi.org/10.1080/15548627.2015.1100356
Lahiri V, Hawkins WD, Klionsky DJ (2019) Watch what you (Self-) eat: autophagic mechanisms that modulate metabolism. Cell Metab 29(4):803–826. https://doi.org/10.1016/j.cmet.2019.03.003
Li H, Yu Z, Zhang W (2020) Misfolded protein aggregation and altered cellular pathways in neurodegenerative diseases. STEMedicine 1(4):e63. https://doi.org/10.37175/stemedicine.v1i4.63
Xi Y, Dhaliwal JS, Ceizar M, Vaculik M, Kumar KL, Lagace DC (2016) Knockout of Atg5 delays the maturation and reduces the survival of adult-generated neurons in the hippocampus. Cell Death Dis 7:e2127. https://doi.org/10.1038/cddis.2015.406
Nowosad A, Besson A (2022) Lysosomes at the crossroads of cell metabolism, cell cycle, and stemness. Int J Mol Sci 23(4):2290. https://doi.org/10.3390/ijms23042290
Jin M, Klionsky DJ (2014) Regulation of autophagy: modulation of the size and number of autophagosomes. FEBS Lett 588(15):2457–2463. https://doi.org/10.1016/j.febslet.2014.06.015
Webber JL, Tooze SA (2010) New insights into the function of Atg9. FEBS Lett 584(7):1319–1326. https://doi.org/10.1016/j.febslet.2010.01.020
Mizushima N, Yoshimori T, Ohsumi Y (2011) The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol 27:107–132. https://doi.org/10.1146/annurev-cellbio-092910-154005
Yamamoto H, Kakuta S, Watanabe TM, Kitamura A, Sekito T, Kondo-Kakuta C, Ichikawa R, Kinjo M, Ohsumi Y (2012) Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J Cell Biol 198(2):219–233. https://doi.org/10.1083/jcb.201202061
Mari M, Griffith J, Rieter E, Krishnappa L, Klionsky DJ, Reggiori F (2010) An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J Cell Biol 190(6):1005–1022. https://doi.org/10.1083/jcb.200912089
Popovic D, Dikic I (2014) TBC1D5 and the AP2 complex regulate ATG9 trafficking and initiation of autophagy. EMBO Rep 15(4):392–401. https://doi.org/10.1002/embr.201337995
Tamura H, Shibata M, Koike M, Sasaki M, Uchiyama Y (2010) Atg9A protein, an autophagy-related membrane protein, is localized in the neurons of mouse brains. J Histochem Cytochem: Off J Histochem Soc 58(5):443–453. https://doi.org/10.1369/jhc.2010.955690
Wen JK, Wang YT, Chan CC, Hsieh CW, Liao HM, Hung CC, Chen GC (2017) Atg9 antagonizes TOR signaling to regulate intestinal cell growth and epithelial homeostasis in Drosophila. eLife. https://doi.org/10.7554/eLife.29338
Hietakangas V, Cohen SM (2009) Regulation of tissue growth through nutrient sensing. Annu Rev Genet 43:389–410. https://doi.org/10.1146/annurev-genet-102108-134815
Zhang W, Thompson BJ, Hietakangas V, Cohen SM (2011) MAPK/ERK signaling regulates insulin sensitivity to control glucose metabolism in Drosophila. PLoS Genet 7(12):e1002429. https://doi.org/10.1371/journal.pgen.1002429
Liu Y, Mattila J, Ventela S, Yadav L, Zhang W, Lamichane N, Sundstrom J, Kauko O, Grenman R, Varjosalo M, Westermarck J, Hietakangas V (2017) PWP1 mediates nutrient-dependent growth control through nucleolar regulation of ribosomal gene expression. Dev Cell 43(2):240–252.e245. https://doi.org/10.1016/j.devcel.2017.09.022
Ma T, Hoeffer CA, Capetillo-Zarate E, Yu F, Wong H, Lin MT, Tampellini D, Klann E, Blitzer RD, Gouras GK (2010) Dysregulation of the mTOR pathway mediates impairment of synaptic plasticity in a mouse model of Alzheimer’s disease. PloS One 5(9):e12845. https://doi.org/10.1371/journal.pone.0012845
Martina JA, Chen Y, Gucek M, Puertollano R (2012) MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy 8(6):903–914. https://doi.org/10.4161/auto.19653
Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, Guan JL, Oshiro N, Mizushima N (2009) Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell 20(7):1981–1991. https://doi.org/10.1091/mbc.E08-12-1248
Wei Y, Xu X (2016) UFMylation: a unique & fashionable modification for life. Genom Proteom Bioinform 14(3):140–146. https://doi.org/10.1016/j.gpb.2016.04.001
Gerakis Y, Quintero M, Li H, Hetz C (2019) The UFMylation system in proteostasis and beyond. Trends Cell Biol 29(12):974–986. https://doi.org/10.1016/j.tcb.2019.09.005
Cai Y, Singh N, Li H (2016) Essential role of Ufm1 conjugation in the hematopoietic system. Exp Hematol 44(6):442–446. https://doi.org/10.1016/j.exphem.2016.03.007
Walczak CP, Leto DE, Zhang L, Riepe C, Muller RY, DaRosa PA, Ingolia NT, Elias JE, Kopito RR (2019) Ribosomal protein RPL26 is the principal target of UFMylation. Proc Natl Acad Sci USA. https://doi.org/10.1073/pnas.1816202116
Yoo HM, Kang SH, Kim JY, Lee JE, Seong MW, Lee SW, Ka SH, Sou YS, Komatsu M, Tanaka K, Lee ST, Noh DY, Baek SH, Jeon YJ, Chung CH (2014) Modification of ASC1 by UFM1 is crucial for ERalpha transactivation and breast cancer development. Mol Cell 56(2):261–274. https://doi.org/10.1016/j.molcel.2014.08.007
Qin B, Yu J, Nowsheen S, Wang M, Tu X, Liu T, Li H, Wang L, Lou Z (2019) UFL1 promotes histone H4 Ufmylation and ATM activation. Nat Commun 10(1):1242. https://doi.org/10.1038/s41467-019-09175-0
Duan R, Shi Y, Yu L, Zhang G, Li J, Lin Y, Guo J, Wang J, Shen L, Jiang H, Wang G, Tang B (2016) UBA5 mutations cause a new form of autosomal recessive cerebellar ataxia. PloS One 11(2):e0149039. https://doi.org/10.1371/journal.pone.0149039
Liu J, Guan D, Dong M, Yang J, Wei H, Liang Q, Song L, Xu L, Bai J, Liu C, Mao J, Zhang Q, Zhou J, Wu X, Wang M, Cong YS (2020) UFMylation maintains tumour suppressor p53 stability by antagonizing its ubiquitination. Nat Cell Biol 22(9):1056–1063. https://doi.org/10.1038/s41556-020-0559-z
Verma P, Augustine GJ, Ammar MR, Tashiro A, Cohen SM (2015) A neuroprotective role for microRNA miR-1000 mediated by limiting glutamate excitotoxicity. Nat Neurosci 18(3):379–385. https://doi.org/10.1038/nn.3935
Cai Y, Pi W, Sivaprakasam S, Zhu X, Zhang M, Chen J, Makala L, Lu C, Wu J, Teng Y, Pace B, Tuan D, Singh N, Li H (2015) UFBP1, a key component of the Ufm1 conjugation system, is essential for ufmylation-mediated regulation of erythroid development. PLoS Genet 11(11):e1005643. https://doi.org/10.1371/journal.pgen.1005643
Chen F, Xing C, Zhang W, Li J, Hu T, Li L, Li H, Cai Y (2019) Salubrinal, a novel inhibitor of eIF-2alpha dephosphorylation, promotes erythropoiesis at early stage targeted by Ufmylation pathway. J Cell Physiol 234(10):18560–18570. https://doi.org/10.1002/jcp.28493
Zhu Y, Lei Q, Li D, Zhang Y, Jiang X, Hu Z, Xu G (2018) Proteomic and biochemical analyses reveal a novel mechanism for promoting protein ubiquitination and degradation by UFBP1, a key component of ufmylation. J Proteome Res 17(4):1509–1520. https://doi.org/10.1021/acs.jproteome.7b00843
Rubinsztein DC, Shpilka T, Elazar Z (2012) Mechanisms of autophagosome biogenesis. Curr Biol: CB 22(1):R29-34. https://doi.org/10.1016/j.cub.2011.11.034
Ravikumar B, Sarkar S, Davies JE, Futter M, Garcia-Arencibia M, Green-Thompson ZW, Jimenez-Sanchez M, Korolchuk VI, Lichtenberg M, Luo S, Massey DC, Menzies FM, Moreau K, Narayanan U, Renna M, Siddiqi FH, Underwood BR, Winslow AR, Rubinsztein DC (2010) Regulation of mammalian autophagy in physiology and pathophysiology. Physiol Rev 90(4):1383–1435. https://doi.org/10.1152/physrev.00030.2009
Bartlett BJ, Isakson P, Lewerenz J, Sanchez H, Kotzebue RW, Cumming RC, Harris GL, Nezis IP, Schubert DR, Simonsen A, Finley KD (2011) p62, Ref(2)P and ubiquitinated proteins are conserved markers of neuronal aging, aggregate formation and progressive autophagic defects. Autophagy 7(6):572–583. https://doi.org/10.4161/auto.7.6.14943
Branon TC, Bosch JA, Sanchez AD, Udeshi ND, Svinkina T, Carr SA, Feldman JL, Perrimon N, Ting AY (2018) Efficient proximity labeling in living cells and organisms with TurboID. Nat Biotechnol 36(9):880–887. https://doi.org/10.1038/nbt.4201
May DG, Scott KL, Campos AR, Roux KJ (2020) Comparative application of BioID and TurboID for protein-proximity biotinylation. Cells 9(5):1070. https://doi.org/10.3390/cells9051070
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2(5):401–404. https://doi.org/10.1158/2159-8290.CD-12-0095
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6(269):p11. https://doi.org/10.1126/scisignal.2004088
Cao Y, Li R, Shen M, Li C, Zou Y, Jiang Q, Liu S, Lu C, Li H, Liu H, Cai Y (2021) DDRGK1, a crucial player of Ufmylation system, is indispensable for autophagic degradation by regulating lysosomal function. Cell Death Dis 12(5):416. https://doi.org/10.1038/s41419-021-03694-9
Schleich S, Teleman AA (2009) Akt phosphorylates both Tsc1 and Tsc2 in Drosophila, but neither phosphorylation is required for normal animal growth. PloS one 4(7):e6305. https://doi.org/10.1371/journal.pone.0006305
Ziviani E, Tao RN, Whitworth AJ (2010) Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. Proc Natl Acad Sci USA 107(11):5018–5023. https://doi.org/10.1073/pnas.0913485107
Cisneros J, Belton TB, Shum GC, Molakal CG, Wong YC (2022) Mitochondria-lysosome contact site dynamics and misregulation in neurodegenerative diseases. Trends Neurosci 45(4):312–322. https://doi.org/10.1016/j.tins.2022.01.005
El Fissi N, Rojo M, Aouane A, Karatas E, Poliacikova G, David C, Royet J, Rival T (2018) Mitofusin gain and loss of function drive pathogenesis in Drosophila models of CMT2A neuropathy. EMBO Rep. https://doi.org/10.15252/embr.201745241
Aparicio R, Rana A, Walker DW (2019) Upregulation of the autophagy adaptor p62/SQSTM1 prolongs health and lifespan in middle-aged Drosophila. Cell Rep 28(4):1029-1040.e1025. https://doi.org/10.1016/j.celrep.2019.06.070
Ramachandran A, Jaeschke H (2019) Acetaminophen hepatotoxicity. Semin Liver Dis 39(2):221–234. https://doi.org/10.1055/s-0039-1679919
Win S, Than TA, Kaplowitz N (2018) The regulation of JNK signaling pathways in cell death through the interplay with mitochondrial SAB and upstream post-translational effects. Int J Mol Sci 19(11):3657. https://doi.org/10.3390/ijms19113657
Biteau B, Karpac J, Hwangbo D, Jasper H (2011) Regulation of Drosophila lifespan by JNK signaling. Exp Gerontol 46(5):349–354. https://doi.org/10.1016/j.exger.2010.11.003
Martin-Blanco E, Gampel A, Ring J, Virdee K, Kirov N, Tolkovsky AM, Martinez-Arias A (1998) Puckered encodes a phosphatase that mediates a feedback loop regulating JNK activity during dorsal closure in Drosophila. Genes Dev 12(4):557–570. https://doi.org/10.1101/gad.12.4.557
Nalls MA et al (2014) Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat Genet 46(9):989–993. https://doi.org/10.1038/ng.3043
Kinghorn KJ, Gronke S, Castillo-Quan JI, Woodling NS, Li L, Sirka E, Gegg M, Mills K, Hardy J, Bjedov I, Partridge L (2016) A Drosophila model of neuronopathic gaucher disease demonstrates lysosomal-autophagic defects and altered mtor signalling and is functionally rescued by rapamycin. J Neurosci: Off J Soc Neurosci 36(46):11654–11670. https://doi.org/10.1523/JNEUROSCI.4527-15.2016
Senturk M, Lin G, Zuo Z, Mao D, Watson E, Mikos AG, Bellen HJ (2019) Ubiquilins regulate autophagic flux through mTOR signalling and lysosomal acidification. Nat Cell Biol 21(3):384–396. https://doi.org/10.1038/s41556-019-0281-x
Kim M, Sandford E, Gatica D, Qiu Y, Liu X, Zheng Y, Schulman BA, Xu J, Semple I, Ro SH, Kim B, Mavioglu RN, Tolun A, Jipa A, Takats S, Karpati M, Li JZ, Yapici Z, Juhasz G, Lee JH, Klionsky DJ, Burmeister M (2016) Mutation in ATG5 reduces autophagy and leads to ataxia with developmental delay. eLife. https://doi.org/10.7554/eLife.12245
Juhasz G, Erdi B, Sass M, Neufeld TP (2007) Atg7-dependent autophagy promotes neuronal health, stress tolerance, and longevity but is dispensable for metamorphosis in Drosophila. Genes Dev 21(23):3061–3066. https://doi.org/10.1101/gad.1600707
Kim M, Park HL, Park HW, Ro SH, Nam SG, Reed JM, Guan JL, Lee JH (2013) Drosophila Fip200 is an essential regulator of autophagy that attenuates both growth and aging. Autophagy 9(8):1201–1213. https://doi.org/10.4161/auto.24811
Cheng Y, Niu Z, Cai Y, Zhang W (2022) Emerging role of UFMylation in secretory cells involved in the endocrine system by maintaining ER proteostasis. Front Endocrinol 13:1085408. https://doi.org/10.3389/fendo.2022.1085408
Wang Z, Gong Y, Peng B, Shi R, Fan D, Zhao H, Zhu M, Zhang H, Lou Z, Zhou J, Zhu WG, Cong YS, Xu X (2019) MRE11 UFMylation promotes ATM activation. Nucleic Acids Res 47(8):4124–4135. https://doi.org/10.1093/nar/gkz110
Xu P, Damschroder D, Zhang M, Ryall KA, Adler PN, Saucerman JJ, Wessells RJ, Yan Z (2019) Atg2, Atg9 and Atg18 in mitochondrial integrity, cardiac function and healthspan in Drosophila. J Mol Cell Cardiol 127:116–124. https://doi.org/10.1016/j.yjmcc.2018.12.006
Stern M, McNew JA (2021) A transition to degeneration triggered by oxidative stress in degenerative disorders. Mol Psychiatry 26(3):736–746. https://doi.org/10.1038/s41380-020-00943-9
Tang HW, Liao HM, Peng WH, Lin HR, Chen CH, Chen GC (2013) Atg9 interacts with dTRAF2/TRAF6 to regulate oxidative stress-induced JNK activation and autophagy induction. Dev Cell 27(5):489–503. https://doi.org/10.1016/j.devcel.2013.10.017
Mailler E, Guardia CM, Bai X, Jarnik M, Williamson CD, Li Y, Maio N, Golden A, Bonifacino JS (2021) The autophagy protein ATG9A enables lipid mobilization from lipid droplets. Nat Commun 12(1):6750. https://doi.org/10.1038/s41467-021-26999-x
Matoba K, Kotani T, Tsutsumi A, Tsuji T, Mori T, Noshiro D, Sugita Y, Nomura N, Iwata S, Ohsumi Y, Fujimoto T, Nakatogawa H, Kikkawa M, Noda NN (2020) Atg9 is a lipid scramblase that mediates autophagosomal membrane expansion. Nat Struct Mol Biol 27(12):1185–1193. https://doi.org/10.1038/s41594-020-00518-w
Maeda S, Yamamoto H, Kinch LN, Garza CM, Takahashi S, Otomo C, Grishin NV, Forli S, Mizushima N, Otomo T (2020) Structure, lipid scrambling activity and role in autophagosome formation of ATG9A. Nat Struct Mol Biol 27(12):1194–1201. https://doi.org/10.1038/s41594-020-00520-2
Campisi D, Desrues L, Dembele KP, Mutel A, Parment R, Gandolfo P, Castel H, Morin F (2022) The core autophagy protein ATG9A controls dynamics of cell protrusions and directed migration. J Cell Biol. https://doi.org/10.1083/jcb.202106014
Kiss V, Jipa A, Varga K, Takats S, Maruzs T, Lorincz P, Simon-Vecsei Z, Szikora S, Foldi I, Bajusz C, Toth D, Vilmos P, Gaspar I, Ronchi P, Mihaly J, Juhasz G (2020) Drosophila Atg9 regulates the actin cytoskeleton via interactions with profilin and Ena. Cell Death Differ 27(5):1677–1692. https://doi.org/10.1038/s41418-019-0452-0
Soukup SF, Kuenen S, Vanhauwaert R, Manetsberger J, Hernandez-Diaz S, Swerts J, Schoovaerts N, Vilain S, Gounko NV, Vints K, Geens A, De Strooper B, Verstreken P (2016) A LRRK2-dependent EndophilinA phosphoswitch is critical for macroautophagy at presynaptic terminals. Neuron 92(4):829–844. https://doi.org/10.1016/j.neuron.2016.09.037
Acknowledgements
We thank Dr. Tong Chao (Zhejiang University) for kindly providing UAS-GFP-Atg9 flies. We also thank Dr. Chen Guang-Chao (National Taiwan University) and Dr. Chen Yawen for providing pUAST-Flag-Atg9 plasmid. We thank Dr. Cai Yu (Temasek Life Sciences laboratory, Singapore) for pActin-Gal4 plasmid.
Funding
This work was supported by the National Natural Science Foundation of China (No. 31701229, 32270516 and 31970413), Senior Health Research Project of Jiangsu Provincial Health Commission (LKM2023010) and National Key R&D Program of China (2018YFC1200201).
Author information
Authors and Affiliations
Contributions
WZ designed the experiments. HL, ZY, ZN, YC, ZW, LH and JZ performed the experiments. HL, ZY, ZN, YC, YC, FM and ZW analyzed the data. WZ, ZY and YC wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Ethical approval and consent to participate
Not applicable.
Consent for publication
All authors read and approved the final manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, H., Yu, Z., Niu, Z. et al. A neuroprotective role of Ufmylation through Atg9 in the aging brain of Drosophila. Cell. Mol. Life Sci. 80, 129 (2023). https://doi.org/10.1007/s00018-023-04778-9
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-023-04778-9