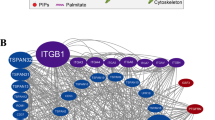
Abstract
EWI2 is a transmembrane immunoglobulin superfamily (IgSF) protein that physically associates with tetraspanins and integrins. It inhibits cancer cells by influencing the interactions among membrane molecules including the tetraspanins and integrins. The present study revealed that, upon EWI2 silencing or ablation, the elevated movement and proliferation of cancer cells in vitro and increased cancer metastatic potential and malignancy in vivo are associated with (i) increases in clustering, endocytosis, and then activation of EGFR and (ii) enhancement of Erk MAP kinase signaling. These changes in signaling make cancer cells (i) undergo partial epithelial-to-mesenchymal (EMT) for more tumor progression and (ii) proliferate faster for better tumor formation. Inhibition of EGFR or Erk kinase can abrogate the cancer cell phenotypes resulting from EWI2 removal. Thus, to inhibit cancer cells, EWI2 prevents EGFR from clustering and endocytosis to restrain its activation and signaling.








Similar content being viewed by others
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Abbreviations
- Ab:
-
Antibody
- BSA:
-
Bovine serum albumin
- CAMs:
-
Cell adhesion molecules
- CHC:
-
Clathrin heavy chain
- ECM:
-
Extracellular matrix
- EGFR:
-
Epidermal growth factor receptor
- EMT:
-
Epithelial-to-mesenchymal transition
- FBS:
-
Fetal bovine serum
- FN:
-
Fibronectin
- HB-EGF:
-
Heparin-binding EGF-like growth factor
- IgSF:
-
Immunoglobulin superfamily
- KO:
-
Knockout
- KD:
-
Knockdown
- LN:
-
Laminin
- mAb:
-
Monoclonal antibody
- MAPK:
-
Mitogen-activated protein kinase
- pAb:
-
Polyclonal antibody
- PBS:
-
Phosphate buffered saline
- PFA:
-
Paraformaldehyde
- PRAD:
-
Prostate adenocarcinoma
- ROI:
-
Region of interest
- STORM:
-
Stochastic optical reconstruction microscopy
- SR:
-
Super-resolution
- TEMD:
-
Tetraspanin-enriched membrane domain
- TUNEL:
-
Terminal deoxynucleotidyl transferase dUTP nick end labeling
References
Charrin S, Le Naour F, Labas V et al (2003) EWI-2 is a new component of the tetraspanin web in hepatocytes and lymphoid cells. Biochem J 373(Pt 2):409–421
Stipp CS, Kolesnikova TV, Hemler ME (2001) EWI-2 is a major CD9 and CD81 partner and member of a novel Ig protein subfamily. J Biol Chem 276(44):40545–40554
Zhang XA, Lane WS, Charrin S et al (2003) EWI2/PGRL associates with the metastasis suppressor KAI1/CD82 and inhibits the migration of prostate cancer cells. Cancer Res 63(10):2665–2674
Charrin S, Jouannet S, Boucheix C et al (2014) Tetraspanins at a glance. J Cell Sci 127(Pt 17):3641–3648
Wang H-X, Li Q, Sharma C et al (2011) Tetraspanin protein contributions to cancer. Biochem Soc Trans 39(2):547–552
Yáñez-Mó M, Barreiro O, Gordon-Alonso M et al (2009) Tetraspanin-enriched microdomains: a functional unit in cell plasma membranes. Trends Cell Biol 19(9):434–446
Yang Y-G, Sari IN, Zia MF et al (2016) Tetraspanins: spanning from solid tumors to hematologic malignancies. Exp Hematol 44(5):322–328
Avraham R, Yarden Y (2011) Feedback regulation of EGFR signalling: decision making by early and delayed loops. Nat Rev Mol Cell Biol 12(2):104–117
Berditchevski F, Odintsova E (2016) ErbB receptors and tetraspanins: casting the net wider. Int J Biochem Cell Biol 77(Pt A):68–71
Mitamura T, Iwamoto R, Umata T et al (1992) The 27-kD diphtheria toxin receptor-associated protein (DRAP27) from vero cells is the monkey homologue of human CD9 antigen: expression of DRAP27 elevates the number of diphtheria toxin receptors on toxin-sensitive cells. J Cell Biol 118(6):1389–1399
Tang M, Yin G, Wang F et al (2015) Downregulation of CD9 promotes pancreatic cancer growth and metastasis through upregulation of epidermal growth factor on the cell surface. Oncol Rep 34(1):350–358
Wang G-P, Han X-F (2015) CD9 modulates proliferation of human glioblastoma cells via epidermal growth factor receptor signaling. Mol Med Rep 12(1):1381–1386
Murayama Y, Shinomura Y, Oritani K et al (2008) The tetraspanin CD9 modulates epidermal growth factor receptor signaling in cancer cells. J Cell Physiol 216(1):135–143
Haeger A, Krause M, Wolf K et al (2014) Cell jamming: collective invasion of mesenchymal tumor cells imposed by tissue confinement. Biochim Biophys Acta 1840(8):2386–2395
Wolf K, Friedl P (2011) Extracellular matrix determinants of proteolytic and non-proteolytic cell migration. Trends Cell Biol 21(12):736–744
Taddei ML, Giannoni E, Comito G et al (2013) Microenvironment and tumor cell plasticity: an easy way out. Cancer Lett 341(1):80–96
Tester AM, Ruangpanit N, Anderson RL et al (2000) MMP-9 secretion and MMP-2 activation distinguish invasive and metastatic sublines of a mouse mammary carcinoma system showing epithelial-mesenchymal transition traits. Clin Exp Metastasis 18(7):553–560
Kong D, Wang Z, Sarkar SH et al (2008) Platelet-derived growth factor-D overexpression contributes to epithelial-mesenchymal transition of PC3 prostate cancer cells. Stem Cells 26(6):1425–1435
Sun Y, Schaar A, Sukumaran P et al (2018) TGFβ-induced epithelial-to-mesenchymal transition in prostate cancer cells is mediated via TRPM7 expression. Mol Carcinog 57(6):752–761
Javadi S, Zhiani M, Mousavi MA et al (2020) Crosstalk between epidermal growth factor receptors (EGFR) and integrins in resistance to EGFR tyrosine kinase inhibitors (TKIs) in solid tumors. Eur J Cell Biol 99(4):151083
Montanari M, Rossetti S, Cavaliere C et al (2017) Epithelial-mesenchymal transition in prostate cancer: an overview. Oncotarget 8(21):35376–35389
Sala-Valdés M, Ursa A, Charrin S et al (2006) EWI-2 and EWI-F link the tetraspanin web to the actin cytoskeleton through their direct association with ezrin-radixin-moesin proteins. J Biol Chem 281(28):19665–19675
Smith CS, Joseph N, Rieger B et al (2010) Fast, single-molecule localization that achieves theoretically minimum uncertainty. Nat Methods 7(5):373–375
Huang F, Schwartz SL, Byars JM et al (2011) Simultaneous multiple-emitter fitting for single molecule super-resolution imaging. Biomed Opt Express 2(5):1377–1393
Ester M, Kriegel H-P, Sander J et al (1996) A density-based algorithm for discovering clusters in large spatial databases with noise. In: Proceedings of the Second International Conference on Knowledge Discovery and Data Mining. Portland, OR, USA, pp 226–31
Daszykowski M, Walczak B, Massart DL et al (2002) Looking for natural patterns in analytical data. 2. Tracing local density with OPTICS. J Chem Inf Comput Sci 42(3):500–507
Johnson S, Chen H, Lo P-K (2013) In vitro tumorsphere formation assays. Bio Protoc 3(3):e325
Portillo-Lara R, Alvarez MM (2015) Enrichment of the cancer stem phenotype in sphere cultures of prostate cancer cell lines occurs through activation of developmental pathways mediated by the transcriptional regulator ΔNp63α. PLoS ONE 10(6):e0130118
He B, Zhang YH, Richardson MM et al (2011) Differential functions of phospholipid binding and palmitoylation of tumour suppressor EWI2/PGRL. Biochem J 437(3):399–411
Li S, Goncalves KA, Lyu B et al (2020) Chemosensitization of prostate cancer stem cells in mice by angiogenin and plexin-B2 inhibitors. Commun Biol. 3(1):26
Collins AT, Berry PA, Hyde C et al (2005) Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res 65(23):10946–10951
Huang C, Fu C, Wren JD et al (2018) Tetraspanin-enriched microdomains regulate digitation junctions. Cell Mol Life Sci 75(18):3423–3439
Noguchi S, Saito A, Nagase T (2018) YAP/TAZ signaling as a molecular link between fibrosis and cancer. Int J Mol Sci 19(11):3674
Zhengming Wu, Guan K-L (2021) Hippo signaling in embryogenesis and development. Trends Biochem Sci 46(1):51–63
Yarden Y, Shilo B-Z (2007) SnapShot: EGFR signaling pathway. Cell 131(5):1018
Sigismund S, Argenzio E, Tosoni D et al (2008) Clathrin-mediated internalization is essential for sustained EGFR signaling but dispensable for degradation. Dev Cell 15(2):209–219
Bakker J, Spits M, Neefjes J et al (2017) The EGFR odyssey—from activation to destruction in space and time. J Cell Sci 130(24):4087–4096
Wang H-X, Sharma C, Knoblich K et al (2015) EWI-2 negatively regulates TGF-β signaling leading to altered melanoma growth and metastasis. Cell Res 25(3):370–385
Yang XH, Kovalenko OV, Kolesnikova TV et al (2006) Contrasting effects of EWI proteins, integrins, and protein palmitoylation on cell surface CD9 organization. J Biol Chem 281(18):12976–12985
Stipp CS (2010) Laminin-binding integrins and their tetraspanin partners as potential antimetastatic targets. Expert Rev Mol Med 12:e3
Gustafson-Wagner E, Stipp CS (2013) The CD9/CD81 tetraspanin complex and tetraspanin CD151 regulate α3β1 integrin-dependent tumor cell behaviors by overlapping but distinct mechanisms. PLoS ONE 8(4):e61834
Hong I-K, Byun H-J, Lee J et al (2014) The tetraspanin CD81 protein increases melanoma cell motility by up-regulating metalloproteinase MT1-MMP expression through the pro-oncogenic Akt-dependent Sp1 activation signaling pathways. J Biol Chem 289(22):15691–15704
Kotha J, Longhurst C, Appling W et al (2008) Tetraspanin CD9 regulates beta 1 integrin activation and enhances cell motility to fibronectin via a PI-3 kinase-dependent pathway. Exp Cell Res 314(8):1811–1822
Posor Y, Eichhorn-Grünig M, Haucke V (2015) Phosphoinositides in endocytosis. Biochim Biophys Acta 1851(6):794–804
Charrin S, Manié S, Thiele C et al (2003) A physical and functional link between cholesterol and tetraspanins. Eur J Immunol 33(9):2479–2489
Zimmerman B, Kelly B, McMillan BJ et al (2016) Cell 167(4):1041-1051.e11
Weber GF, Bjerke MA, DeSimone DW (2011) Integrins and cadherins join forces to form adhesive networks. J Cell Sci 124(Pt 8):1183–1193
Mui KL, Chen CS, Assoian RK (2016) The mechanical regulation of integrin-cadherin crosstalk organizes cells, signaling and forces. J Cell Sci 129(6):1093–1100
Rausch S, Das T, Soiné JRD et al (2013) Polarizing cytoskeletal tension to induce leader cell formation during collective cell migration. Biointerphases 8(1):32
Chenying Fu, Zhang Q, Wang A et al (2021) EWI-2 controls nucleocytoplasmic shuttling of EGFR signaling molecules and miRNA sorting in exosomes to inhibit prostate cancer cell metastasis. Mol Oncol 15(5):1543–1565
Hill MM, Bastiani M, Luetterforst R et al (2008) PTRF-Cavin, a conserved cytoplasmic protein required for caveola formation and function. Cell 132(1):113–124
Peng Wu, Wee P, Jiang J et al (2012) Differential regulation of transcription factors by location-specific EGF receptor signaling via a spatio-temporal interplay of ERK activation. PLoS ONE 7(9):e41354
Gayer CP, Craig DH, Flanigan TL et al (2010) ERK regulates strain-induced migration and proliferation from different subcellular locations. J Cell Biochem 109(4):711–725
Carraway KL 3rd, Sweeney C (2006) Co-opted integrin signaling in ErbB2-induced mammary tumor progression. Cancer Cell 10(2):93–95
Alexi X, Berditchevski F, Odintsova E (2011) The effect of cell-ECM adhesion on signalling via the ErbB family of growth factor receptors. Biochem Soc Trans 39(2):568–573
Erfani S, Hua H, Pan Y et al (2021) The context-dependent impact of integrin-associated CD151 and other tetraspanins on cancer development and progression: a class of versatile mediators of cellular function and signaling, tumorigenesis and metastasis. Cancers (Basel) 13(9):2005
Ramovs V, Te Molder L, Sonnenberg A (2017) The opposing roles of laminin-binding integrins in cancer. Matrix Biol 57–58:213–243
Acknowledgements
We thank Drs. Felipe V. Catalan and Shoshana Levy of Stanford University for providing EWI2 CRISPR/Cas9 KO system and comments, Ms. Kathy Kyler for English editing, the OMRF imaging facility for image acquisition and analysis, and OUHSC Stephenson Cancer Center tissue pathology core and functional genomics core facility.
Funding
This work was supported by OCAST grants HR13-207 and HR20-055, the research grants from OCASCR (a program of TSET), and the University of Oklahoma Health Science Center to XAZ. XAZ is an Oklahoma TSET Cancer Research Scholar.
Author information
Authors and Affiliations
Contributions
CF, JW, and SP performed experiments, analyzed data, and wrote manuscript. JDW and K-KW analyzed data. YD, JC, and YY performed experiments. HK and MJ provided technical advice. AM and TT provided special reagent and/or technical advice. KAL designed experiments and analyzed data. XAZ designed experiments, analyzed data, and wrote manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing financial interests.
Ethics statement
All procedures involving animals were performed according to protocols approved by the Institutional Animal Care and Use Committee (IACUC).
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
18_2022_4417_MOESM1_ESM.tif
Figure S1 EWI2 KD and KO in PC3 cells. A. EWI2 expression levels at the cell surface of PC3 cells upon EWI2 KD, as analyzed with flow cytometry and presented as MFI (mean±SD, n=3 individual measurements). ** p<0.01. Isotype matched IgG staining serves as a negative control. B. Western blot analysis of EWI2 protein levels in PC3 cells upon EWI2 KD. Actin serves as a protein loading control. C. EWI2 expression levels at the surfaces of PC3 cells upon EWI2 KO were measured by flow cytometry and expressed as MFI (mean±SD, n=3 individual measurements). ** p<0.01. D. Western blot analysis on EWI2 protein levels in PC3 cells upon EWI2 KO. Actin serves as a protein loading control (TIF 802 KB)
18_2022_4417_MOESM2_ESM.tif
Figure S2 Effects of EWI2 removals on the movement of PC3 cells. A. PC3 cells transfected with control siRNA (NEG) or EWI-2 siRNA (KD) were analyzed in Transwell migration and invasion assays. The cells that moved through the insert pores and adhered onto the bottom of the inserts were photographed. Scale bars: 500 µm for Transwell migration on FN, 250 µm for Transwell migration on LN111, and 100 µm for invasion. B. PC3-control (NEG) and -EWI2-null (KO) cells were examined for the migration through Transwell inserts, which were coated with either FN (10 μg/ml) or LN111 (10 μg/ml). The cells that migrated through the insert pores and adhered onto the bottom of the inserts were photographed. Scale bars: 300 µm for Transwell migration on FN and LN111. Scale bars: 200 µm for Transwell migration on LN411. C. Immunohistochemical analyses of Ki67 and cleaved Caspase-3 in primary tumor tissues. Xenografts of PC3 cells from athymic nude mice were dissected, sectioned, and immune-stained with Ki67 and cleaved Caspase-3 Abs. Arrows indicate the cells positive in cleaved caspase-3. Scale bars: 75 µm. D. Immunofluorescence staining of the FN and collagen-IV deposited by the cells on the glass coverslips, as described in Figure 3F, were imaged by fluorescence microscopy and quantified with ImageJ as fluorescence units (mean±SD, n=3 individual experiments, 5 random microscopic fields per experiment). The cells on the glass coverslips were stained with crystal violet and quantified as optical density (mean±SD, n=3 individual experiments). ** p<0.01, *** p<0.001, and **** p<0.0001. Scale bars: 50 µm. E. Tumor tissues sections were stained by Sirius Red. Scale bars: 75 µm (TIF 5990 KB)
18_2022_4417_MOESM3_ESM.tif
Figure S3 Examination of PC3 transfectant cells with CD9 mAbs ALB6 and C9BB. A. Flowstream analysis of PC3-Mock and PC3-EWI2 KO cells with CD9 mAbs ALB6 (for total CD9) and C9BB (for homo-clustered CD9). CD9 levels at the cell surfaces were presented as MFI (mean±SD, n=3 individual experiments). ** p<0.05 and ** p<0.01. B. Flowstream images of the representative PC3 cells immune-stained with CD9 mAbs. Scale bars: 10 µm (TIF 766 KB)
18_2022_4417_MOESM4_ESM.tif
Figure S4 Colocalizations of CD9 with clathrin heavy chain and caveolin-1 in PC3-Mock and -EWI2 KO cells. A and B. Colocalizations of CHC (A) and caveolin-1 (B) with total and homo-clustered CD9, stained by CD9 mAbs ALB6 and C9BB, respectively, were examined in immunofluorescence, imaged with confocal microscopy, and quantified as Manders colocalization coefficients (mean±SD, n=3 individual experiments, nine cells per experiment). M1=CHC or caveolin-1-colocalized CD9/CD9, and M2 CD9-colocalized CHC or caveolin-1/CHC or caveolin-1. Scale bar: 6 µm (TIF 5778 KB)
18_2022_4417_MOESM5_ESM.tif
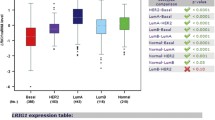
Figure S5 Relationship between EWI2 gene expression and PRAD. A. MEXPRESS (https://mexpress.be/) was used to analyze the relationship between EWI2 mRNA levels and PRAD based on TCGA (https://www.cancer.gov/) data (PRAD: n=617). B. EWI2 alteration in different types of cancers was obtained from cBioPortal (https://www.cbioportal.org/). SKCM: Skin Cutaneous Melanoma. STAD: Stomach adenocarcinoma. C. TCGA data of EWI2 and PRAD were obtained from UALCAN (http://ualcan.path.uab.edu/analysis.html), by grouping them based on sample types (normal: n=52, PRAD: n=497). ****: p<0.0001. D. Kaplan-Meier survival curves for EWI2 in TCGA data were plotted using GEPIA (http://gepia.cancer-pku.cn/) (PRAD: n=246). Patients were divided by median EWI2 expression levels. E. Correlations of EWI2 with EGFR, HER2, and HER3 in gene expression in PRAD were examined in GEPIA (PRAD: n=252). F. Correlations of EWI2 with MEK1, MEK2, Erk1, and Erk2 in gene expression in PRAD were examined in GEPIA (PRAD: n=252) (TIF 1064 KB)
Rights and permissions
About this article
Cite this article
Fu, C., Wang, J., Pallikkuth, S. et al. EWI2 prevents EGFR from clustering and endocytosis to reduce tumor cell movement and proliferation. Cell. Mol. Life Sci. 79, 389 (2022). https://doi.org/10.1007/s00018-022-04417-9
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-022-04417-9