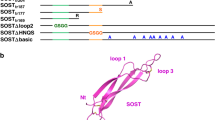
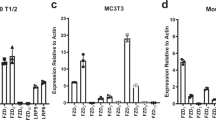
Abstract
Induction of bone formation by Wnt ligands is inhibited when sclerostin (Scl), an osteocyte-produced antagonist, binds to its receptors, the low-density lipoprotein receptor-related proteins 5 or 6 (LRP5/6). Recently, it was shown that enhanced inhibition is achieved by Scl binding to the co-receptor LRP4. However, it is not clear if the binding of Scl to LRP4 facilitates Scl binding to LRP5/6 or inhibits the Wnt pathway in an LRP5/6-independent manner. Here, using the yeast display system, we demonstrate that Scl exhibits a stronger binding affinity for LRP4 than for LRP6. Moreover, we found stronger Scl binding to LRP6 in the presence of LRP4. We further show that a Scl mutant (SclN93A), which tightly binds LRP4 but not LRP6, does not inhibit the Wnt pathway on its own. We demonstrate that SclN93A competes with Scl for a common binding site on LRP4 and antagonizes Scl inhibition of the Wnt signaling pathway in osteoblasts in vitro. Finally, we demonstrate that 2 weeks of bi-weekly subcutaneous injections of SclN93A fused to the fragment crystallizable (Fc) domain of immunoglobulin (SclN93AFc), which retains the antagonistic activity of the mutant, significantly increases bone formation rate and enhances trabecular volumetric bone fraction, trabecular number, and bone length in developing mice. Our data show that LRP4 serves as an anchor that facilitates Scl–LRP6 binding and that inhibition of the Wnt pathway by Scl depends on its prior binding to LRP4. We further provide evidence that compounds that inhibit Scl–LRP4 interactions offer a potential strategy to promote anabolic bone functions.




Similar content being viewed by others
Code availability
Not applicable.
Data availability
Data will be made available on reasonable request.
References
Raggatt LJ, Partridge NC (2010) Cellular and molecular mechanisms of bone remodeling. J Biol Chem. https://doi.org/10.1074/jbc.R109.041087
Robling AG, Castillo AB, Turner CH (2006) Biomechanical and molecular regulation of bone remodeling. Annu Rev Biomed Eng. https://doi.org/10.1146/annurev.bioeng.8.061505.095721
National Institutes of Health (NIH) (2001) National Institutes of Health (NIH) consensus development panel on osteoporosis prevention, diagnosis, and therapy. JAMA
Zeng X et al (2008) Initiation of Wnt signaling: control of Wnt coreceptor Lrp6 phosphorylation/activation via frizzled, dishevelled and axin functions. Development. https://doi.org/10.1242/dev.013540
Khosla S, Westendorf JJ, Oursler MJ (2008) Building bone to reverse osteoporosis and repair fractures. J Clin Investig. https://doi.org/10.1172/JCI33612
Weivoda MM et al (2016) Wnt signaling inhibits osteoclast differentiation by activating canonical and noncanonical cAMP/PKA pathways. J Bone Miner Res. https://doi.org/10.1002/jbmr.2599
Mani A et al (2007) LRP6 mutation in a family with early coronary disease and metabolic risk factors. Science. https://doi.org/10.1126/science.1136370
Kokubu C et al (2004) Skeletal defects in ringelschwanz mutant mice reveal that Lrp6 is required for proper somitogenesis and osteogenesis. Development. https://doi.org/10.1242/dev.01405
Holmen SL et al (2004) Decreased BMD and limb deformities in mice carrying mutations in both Lrp5 and Lrp6. J Bone Miner Res. https://doi.org/10.1359/JBMR.040907
Keupp K et al (2013) Mutations in WNT1 cause different forms of bone fragility. Am J Hum Genet. https://doi.org/10.1016/j.ajhg.2013.02.010
Pyott SM et al (2013) WNT1 mutations in families affected by moderately severe and progressive recessive osteogenesis imperfecta. Am J Hum Genet. https://doi.org/10.1016/j.ajhg.2013.02.009
Lu Y et al (2018) Novel WNT1 mutations in children with osteogenesis imperfecta: clinical and functional characterization. Bone. https://doi.org/10.1016/j.bone.2018.06.018
Zheng HF et al (2012) WNT16 influences bone mineral density, cortical bone thickness, bone strength, and osteoporotic fracture risk. PLoS Genet. https://doi.org/10.1371/journal.pgen.1002745
Medina-Gomez C et al (2012) Meta-analysis of genome-wide scans for total body BMD in children and adults reveals allelic heterogeneity and age-specific effects at the WNT16 locus. PLoS Genet. https://doi.org/10.1371/journal.pgen.1002718
García-Ibarbia C et al (2013) Missense polymorphisms of the WNT16 gene are associated with bone mass, hip geometry and fractures. Osteoporos Int. https://doi.org/10.1007/s00198-013-2302-0
Takada I et al (2007) A histone lysine methyltransferase activated by non-canonical Wnt signalling suppresses PPAR-γ transactivation. Nat Cell Biol. https://doi.org/10.1038/ncb1647
Zhou H et al (2009) Glucocorticoid-dependent Wnt signaling by mature osteoblasts is a key regulator of cranial skeletal development in mice. Development. https://doi.org/10.1242/dev.027706
Tsukamoto M et al (2019) Findings as a starting point to unravel the underlying mechanisms of in vivo interactions involving Wnt10a in bone, fat and muscle. Bone. https://doi.org/10.1016/j.bone.2018.10.009
Laine CM et al (2011) Novel mutations affecting LRP5 splicing in patients with osteoporosis-pseudoglioma syndrome (OPPG). Eur J Hum Genet. https://doi.org/10.1038/ejhg.2011.42
Gong Y et al (2001) LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell
Hartikka H et al (2005) Heterozygous mutations in the LDL receptor-related protein 5 (LRP5) gene are associated with primary osteoporosis in children. J Bone Miner Res. https://doi.org/10.1359/JBMR.050101
Korvala J et al (2012) Mutations in LRP5 cause primary osteoporosis without features of OI by reducing Wnt signaling activity. BMC Med Genet. https://doi.org/10.1186/1471-2350-13-26
Boyden LM et al (2002) High bone density due to a mutation in LDL-receptor-related protein 5. N Engl J Med. https://doi.org/10.1056/NEJMoa013444
Roetzer KM et al (2018) Novel familial mutation of LRP5 causing high bone mass: genetic analysis, clinical presentation, and characterization of bone matrix mineralization. Bone. https://doi.org/10.1016/j.bone.2017.12.002
Whyte MP et al (2019) New explanation for autosomal dominant high bone mass: mutation of low-density lipoprotein receptor-related protein 6. Bone. https://doi.org/10.1016/j.bone.2019.05.003
Winkler DG et al (2003) Osteocyte control of bone formation via sclerostin, a novel BMP antagonist. EMBO J. https://doi.org/10.1093/emboj/cdg599
Brunkow ME et al (2001) Bone dysplasia sclerosteosis results from loss of the SOST gene product, a novel cystine knot-containing protein. Am J Hum Genet. https://doi.org/10.1086/318811
Balemans W et al (2001) Increased bone density in sclerosteosis is due to the deficiency of a novel secreted protein (SOST). Hum Mol Genet. https://doi.org/10.1093/hmg/10.5.537
Balemans W et al (2002) Identification of a 52 kb deletion downstream of the SOST gene in patients with van Buchem disease. J Med Genet. https://doi.org/10.1136/jmg.39.2.91
Semënov M, Tamai K, He X (2005) SOST is a ligand for LRP5/LRP6 and a Wnt signaling inhibitor. J Biol Chem. https://doi.org/10.1074/jbc.M504308200
Li X et al (2005) Sclerostin binds to LRP5/6 and antagonizes canonical Wnt signaling. J Biol Chem. https://doi.org/10.1074/jbc.M413274200
Leupin O et al (2011) Bone overgrowth-associated mutations in the LRP4 gene impair sclerostin facilitator function. J Biol Chem. https://doi.org/10.1074/jbc.M110.190330
Fijalkowski I et al (2016) A novel domain-specific mutation in a sclerosteosis patient suggests a role of LRP4 as an anchor for sclerostin in human bone. J Bone Miner Res. https://doi.org/10.1002/jbmr.2782
Boudin E et al (2017) The Lrp4R1170Q homozygous knock-in mouse recapitulates the bone phenotype of sclerosteosis in humans. J Bone Miner Res. https://doi.org/10.1002/jbmr.3160
Bullock WA et al (2019) Lrp4 mediates bone homeostasis and mechanotransduction through interaction with sclerostin in vivo. iScience. https://doi.org/10.1016/j.isci.2019.09.023
Strickland DK, Gonias SL, Argraves WS (2002) Diverse roles for the LDL receptor family. Trends Endocrinol Metab. https://doi.org/10.1016/S1043-2760(01)00526-4
Bourhis E et al (2011) Wnt antagonists bind through a short peptide to the first β-propeller domain of LRP5/6. Structure. https://doi.org/10.1016/j.str.2011.07.005
Kim J et al (2020) Sclerostin inhibits Wnt signaling through tandem interaction with two LRP6 ectodomains. Nat Commun. https://doi.org/10.1038/s41467-020-19155-4
Veverka V et al (2009) Characterization of the structural features and interactions of sclerostin. Molecular insight into a key regulator of Wnt-mediated bone formation. J Biol Chem. https://doi.org/10.1074/jbc.M807994200
Holdsworth G et al (2012) Characterization of the interaction of sclerostin with the low density lipoprotein receptor-related protein (LRP) family of wnt co-receptors. J Biol Chem. https://doi.org/10.1074/jbc.M112.350108
Boschert V et al (2013) Mutational analysis of sclerostin shows importance of the flexible loop and the cystine-knot for Wnt-signaling inhibition. PLoS ONE. https://doi.org/10.1371/journal.pone.0081710
Jarmoskaite I, Alsadhan I, Vaidyanathan PP, Herschlag D (2020) How to measure and evaluate binding affinities. Elife 9:1–34. https://doi.org/10.7554/ELIFE.57264
Van Deventer JA, Kelly RL, Rajan S, Wittrup KD, Sidhu SS (2015) A switchable yeast display/secretion system. Protein Eng Des Sel 28(10):317–325. https://doi.org/10.1093/PROTEIN/GZV043
Zahradník J et al (2021) SARS-CoV-2 RBD in vitro evolution follows contagious mutation spread, yet generates an able infection inhibitor. BioRxiv. https://doi.org/10.1101/2021.01.06.425392
Bowen J, Schneible J, Bacon K, Labar C, Menegatti S, Rao BM (2021) Screening of yeast display libraries of enzymatically treated peptides to discover macrocyclic peptide ligands. Int J Mol Sci 22:1634. https://doi.org/10.3390/IJMS22041634
Goel S, Chin EN, Fakhraldeen SA, Berry SM, Beebe DJ, Alexander CM (2012) Both LRP5 and LRP6 receptors are required to respond to physiological Wnt ligands in mammary epithelial cells and fibroblasts. J Biol Chem. https://doi.org/10.1074/jbc.M112.362137
Cejka D et al (2014) Renal elimination of sclerostin increases with declining kidney function. J Clin Endocrinol Metab. https://doi.org/10.1210/jc.2013-2786
Strohl WR (2015) Fusion proteins for half-life extension of biologics as a strategy to make biobetters. BioDrugs. https://doi.org/10.1007/s40259-015-0133-6
Tu X et al (2012) Sost downregulation and local Wnt signaling are required for the osteogenic response to mechanical loading. Bone. https://doi.org/10.1016/j.bone.2011.10.025
Chang MK et al (2014) Disruption of Lrp4 function by genetic deletion or pharmacological blockade increases bone mass and serum sclerostin levels. Proc Natl Acad Sci U S A. https://doi.org/10.1073/pnas.1413828111
Krause C et al (2010) Distinct modes of inhibition by sclerostin on bone morphogenetic protein and Wnt signaling pathways. J Biol Chem. https://doi.org/10.1074/jbc.M110.153890
Chao G, Lau WL, Hackel BJ, Sazinsky SL, Lippow SM, Wittrup KD (2006) Isolating and engineering human antibodies using yeast surface display. Nat Protoc. https://doi.org/10.1038/nprot.2006.94
Lipovšek D et al (2007) Evolution of an interloop disulfide bond in high-affinity antibody mimics based on fibronectin type III domain and selected by yeast surface display: molecular convergence with single-domain camelid and shark antibodies. J Mol Biol. https://doi.org/10.1016/j.jmb.2007.02.029
Xiong L et al (2015) Lrp4 in osteoblasts suppresses bone formation and promotes osteoclastogenesis and bone resorption. Proc Natl Acad Sci U S A. https://doi.org/10.1073/pnas.1419714112
Wehrli M et al (2000) Arrow encodes an LDL-receptor-related protein essential for Wingless signalling. Nature 407(6803):527–530. https://doi.org/10.1038/35035110
Li X et al (2008) Targeted deletion of the sclerostin gene in mice results in increased bone formation and bone strength. J Bone Miner Res. https://doi.org/10.1359/jbmr.080216
Lin C et al (2009) Sclerostin mediates bone response to mechanical unloading through antagonizing Wnt/β-catenin signaling. J Bone Miner Res. https://doi.org/10.1359/jbmr.090411
Weatherbee SD, Anderson KV, Niswander LA (2006) LDL-receptor-related protein 4 is crucial for formation of the neuromuscular junction. Development 133(24):4993–5000. https://doi.org/10.1242/dev.02696
Zhang B, Luo S, Wang Q, Suzuki T, Xiong WC, Mei L (2008) LRP4 serves as a coreceptor of agrin. Neuron 60(2):285–297. https://doi.org/10.1016/j.neuron.2008.10.006
Barik A et al (2014) LRP4 is critical for neuromuscular junction maintenance. J Neurosci 34(42):13892–13905. https://doi.org/10.1523/JNEUROSCI.1733-14.2014
Gautam M, Noakes P, Moscoso L (2021) F. R.-cell, and undefined 1996, Defective neuromuscular synaptogenesis in agrin-deficient mutant mice. Elsevier, New York. https://www.sciencedirect.com/science/article/pii/S0092867400812532. Accessed 27 Jun 2021
Holdsworth G et al (2018) Dampening of the bone formation response following repeat dosing with sclerostin antibody in mice is associated with up-regulation of Wnt antagonists. Bone. https://doi.org/10.1016/j.bone.2017.11.003
Chan BY et al (2011) Increased chondrocyte sclerostin may protect against cartilage degradation in osteoarthritis. Osteoarthr Cartil. https://doi.org/10.1016/j.joca.2011.04.014
Bouaziz W, Funck-Brentano T, Lin H, Marty C, Hay E, Cohen-Solal M (2014) Lack of sclerostin promotes osteoarthritis by activating canonical and non-canonical WNT pathways. Osteoarthr Cartil. https://doi.org/10.1016/j.joca.2014.02.629
Chang JC et al (2018) SOST/sclerostin improves posttraumatic osteoarthritis and inhibits MMP2/3 expression after injury. J Bone Miner Res. https://doi.org/10.1002/jbmr.3397
Li J et al (2019) SOST deficiency aggravates osteoarthritis in mice by promoting sclerosis of subchondral bone. Biomed Res Int. https://doi.org/10.1155/2019/7623562
Hudson BD et al (2015) SOST inhibits prostate cancer invasion. PLoS ONE. https://doi.org/10.1371/journal.pone.0142058
Desjardins L et al (2014) Uremic toxicity and sclerostin in chronic kidney disease patients. Nephrol Ther. https://doi.org/10.1016/j.nephro.2014.04.002
Novo-Rodríguez C et al (2018) Circulating levels of sclerostin are associated with cardiovascular mortality. PLoS ONE. https://doi.org/10.1371/journal.pone.0199504
Kanbay M et al (2014) Serum sclerostin and adverse outcomes in nondialyzed chronic kidney disease patients. J Clin Endocrinol Metab. https://doi.org/10.1210/jc.2014-2042
Viaene L et al (2013) Sclerostin: another bone-related protein related to all-cause mortality in haemodialysis? Nephrol Dial Transplant. https://doi.org/10.1093/ndt/gft039
Brandenburg VM et al (2013) Relationship between sclerostin and cardiovascular calcification in hemodialysis patients: a cross-sectional study. BMC Nephrol. https://doi.org/10.1186/1471-2369-14-219
Drechsler C et al (2015) High levels of circulating sclerostin are associated with better cardiovascular survival in incident dialysis patients: results from the NECOSAD study. Nephrol Dial Transplant. https://doi.org/10.1093/ndt/gfu301
Rosenfeld L, Shirian J, Zur Y, Levaot N, Shifman JM, Papo N (2015) Combinatorial and computational approaches to identify interactions of macrophage colony-stimulating factor (M-CSF) and its receptor c-FMS. J Biol Chem. https://doi.org/10.1074/jbc.M115.671271
Rueden CT et al (2017) ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinform. https://doi.org/10.1186/s12859-017-1934-z
Dyment NA et al (2016) High-throughput, multi-image cryohistology of mineralized tissues. J Vis Exp 2016(115):54468. https://doi.org/10.3791/54468
Acknowledgements
This work was supported by grants from the ISRAEL SCIENCE FOUNDATION no. 357/18 (NL) and no. 1615/19 (NP).
Funding
This work was supported by grants from the ISRAEL SCIENCE FOUNDATION no. 357/18 (NL) and no. 1615/19 (NP).
Author information
Authors and Affiliations
Contributions
SK, conceptualization, data curation, investigation, methodology, validation, visualization, writing—original draft, writing—review and editing. BC, methodology. CA-D, methodology. NP, conceptualization, formal analysis, funding acquisition, investigation, methodology, resources, supervision, validation, visualization, writing—original draft, writing—review and editing. NL, conceptualization, formal analysis, funding acquisition, investigation, methodology, resources, supervision, validation, visualization, writing—original draft, writing—review and editing.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethics approval
All mouse studies were carried out according to protocols approved by the Ben-Gurion University Committee for the Ethical Care and Use of Animals in Experiments (permit number: IL-04-01-2019(D)).
Consent to participate
Not applicable.
Consent for publication
All authors agree with the submission, and the work has not been published or submitted for publication elsewhere either completely or in part, or in another form or language.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Katchkovsky, S., Chatterjee, B., Abramovitch-Dahan, CV. et al. Competitive blocking of LRP4–sclerostin binding interface strongly promotes bone anabolic functions. Cell. Mol. Life Sci. 79, 113 (2022). https://doi.org/10.1007/s00018-022-04127-2
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-022-04127-2