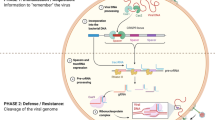
Abstract
Bacteriophage genomes are the richest source of modified nucleobases of any life form. Of these, 2,6 diaminopurine, which pairs with thymine by forming three hydrogen bonds violates Watson and Crick’s base pairing. 2,6 diaminopurine initially found in the cyanophage S-2L is more widespread than expected and has also been detected in phage infecting Gram-negative and Gram-positive bacteria. The biosynthetic pathway for aminoadenine containing DNA as well as the exclusion of adenine are now elucidated. This example of a natural deviation from the genetic code represents only one of the possibilities explored by nature and provides a proof of concept for the synthetic biology of non-canonical nucleic acids.








Similar content being viewed by others
Availability of data and materials
The atomic coordinates of PhiVC8 PurZ, S-2L DatZ and S-2L MazZ have been deposited in the Protein Data Bank with the accession codes 6FLF, 6FM0, 6FM1, 6TNH; 6ZPB and 6ZPC; 7ODY.
References
Carell T, Brandmayr C, Hienzsch A et al (2012) Structure and function of noncanonical nucleobases. Angew Chem Int Ed 51:7110–7131. https://doi.org/10.1002/anie.201201193
Adhikari S, Curtis PD (2016) DNA methyltransferases and epigenetic regulation in bacteria. FEMS Microbiol Rev 40:575–591. https://doi.org/10.1093/femsre/fuw023
Lee Y-J, Dai N, Walsh SE et al (2018) Identification and biosynthesis of thymidine hypermodifications in the genomic DNA of widespread bacterial viruses. Proc Natl Acad Sci USA 115:E3116–E3125. https://doi.org/10.1073/pnas.1714812115
Weigele P, Raleigh EA (2016) Biosynthesis and function of modified bases in bacteria and their viruses. Chem Rev 116:12655–12687. https://doi.org/10.1021/acs.chemrev.6b00114
Lee Y-J, Dai N, Müller SI et al (2021) Pathways of thymidine hypermodification. Nucleic Acids Res. https://doi.org/10.1093/nar/gkab781
Swinton D, Hattman S, Crain PF et al (1983) Purification and characterization of the unusual deoxynucleoside, α-N-(9-beta-d-2’-deoxyribofuranosylpurin-6-yl)glycinamide, specified by the phage Mu modification function. Proc Natl Acad Sci USA 80:7400–7404. https://doi.org/10.1073/pnas.80.24.7400
Thiaville JJ, Kellner SM, Yuan Y et al (2016) Novel genomic island modifies DNA with 7-deazaguanine derivatives. Proc Natl Acad Sci USA 113:E1452-1459. https://doi.org/10.1073/pnas.1518570113
Nikolskaya II, Lopatina NG, Debov SS (1976) Methylated guanine derivative as a minor base in the DNA of phage DDVI Shigella disenteriae. Biochim Biophys Acta 435:206–210. https://doi.org/10.1016/0005-2787(76)90251-3
Dunn DB, Smith JD (1958) The occurrence of 6-methylaminopurine in deoxyribonucleic acids. Biochem J 68:627–636. https://doi.org/10.1042/bj0680627
Crippen CS, Lee Y-J, Hutinet G et al (2019) Deoxyinosine and 7-Deaza-2-deoxyguanosine as carriers of genetic information in the DNA of Campylobacter viruses. J Virol. https://doi.org/10.1128/JVI.01111-19
Hutinet G, Lee Y, de Crécy-Lagard V, Weigele P Hypermodified DNA in Viruses of E. coli and Salmonella. EcoSal Plus 9:eESP-0028–2019
Kirnos MD, Khudyakov IY, Alexandrushkina NI, Vanyushin BF (1977) 2-Aminoadenine is an adenine substituting for a base in S-2L cyanophage DNA. Nature 270:369–370. https://doi.org/10.1038/270369a0
Khudyakov IY, Kirnos MD, Alexandrushkina NI, Vanyushin BF (1978) Cyanophage S-2L contains DNA with 2,6-diaminopurine substituted for adenine. Virology 88:8–18. https://doi.org/10.1016/0042-6822(78)90104-6
Bailly C, Suh D, Waring MJ, Chaires JB (1998) Binding of daunomycin to diaminopurine- and/or inosine-substituted DNA. Biochemistry 37:1033–1045. https://doi.org/10.1021/bi9716128
Bailly C, Waring MJ (1998) The use of diaminopurine to investigate structural properties of nucleic acids and molecular recognition between ligands and DNA. Nucleic Acids Res 26:4309–4314. https://doi.org/10.1093/nar/26.19.4309
Szekeres M, Matveyev AV (1987) Cleavage and sequence recognition of 2,6-diaminopurine-containing DNA by site-specific endonucleases. FEBS Lett 222:89–94. https://doi.org/10.1016/0014-5793(87)80197-7
Honzatko RB, Stayton MM, Fromm HJ (1999) Adenylosuccinate synthetase: recent developments. Adv Enzymol Relat Areas Mol Biol 73(57–102):ix–x
Solís-Sánchez A, Hernández-Chiñas U, Navarro-Ocaña A et al (2016) Genetic characterization of ØVC8 lytic phage for Vibrio cholerae O1. Virol J 13:47. https://doi.org/10.1186/s12985-016-0490-x
Bouyoub A, Barbier G, Forterre P, Labedan B (1996) The adenylosuccinate synthetase from the hyperthermophilic archaeon Pyrococcus species displays unusual structural features. J Mol Biol 261:144–154. https://doi.org/10.1006/jmbi.1996.0448
Jayalakshmi R, Sumathy K, Balaram H (2002) Purification and characterization of recombinant plasmodium falciparum adenylosuccinate synthetase expressed in Escherichia coli. Protein Expr Purif 25:65–72. https://doi.org/10.1006/prep.2001.1610
Powell SM, Zalkin H, Dixon JE (1992) Cloning and characterization of the cDNA encoding human adenylosuccinate synthetase. FEBS Lett 303:4–10. https://doi.org/10.1016/0014-5793(92)80465-S
Honzatko RB, Fromm HJ (1999) Structure-function studies of adenylosuccinate synthetase from Escherichia coli. Arch Biochem Biophys 370:1–8. https://doi.org/10.1006/abbi.1999.1383
Sleiman D, Garcia PS, Lagune M et al (2021) A third purine biosynthetic pathway encoded by aminoadenine-based viral DNA genomes. Science 372:516–520. https://doi.org/10.1126/science.abe6494
Zhou Y, Xu X, Wei Y et al (2021) A widespread pathway for substitution of adenine by diaminopurine in phage genomes. Science 372:512–516. https://doi.org/10.1126/science.abe4882
Iancu CV, Zhou Y, Borza T et al (2006) Cavitation as a mechanism of substrate discrimination by adenylosuccinate synthetases. Biochemistry 45:11703–11711. https://doi.org/10.1021/bi0607498
Kang C, Sun N, Honzatko RB, Fromm HJ (1994) Replacement of Asp333 with Asn by site-directed mutagenesis changes the substrate specificity of Escherichia coli adenylosuccinate synthetase from guanosine 5′-triphosphate to xanthosine 5′-triphosphate. J Biol Chem 269:24046–24049
Pezo V, Jaziri F, Bourguignon P-Y et al (2021) Noncanonical DNA polymerization by aminoadenine-based siphoviruses. Science 372:520–524. https://doi.org/10.1126/science.abe6542
Czernecki D, Hu H, Romoli F, Delarue M (2021) Structural dynamics and determinants of 2-aminoadenine specificity in DNA polymerase DpoZ of vibriophage ϕVC8. Nucleic Acids Res. https://doi.org/10.1093/nar/gkab955
Jeon YJ, Park SC, Song WS et al (2016) Structural and biochemical characterization of bacterial YpgQ protein reveals a metal-dependent nucleotide pyrophosphohydrolase. J Struct Biol 195:113–122. https://doi.org/10.1016/j.jsb.2016.04.002
Vorontsov II, Minasov G, Kiryukhina O et al (2011) Characterization of the deoxynucleotide triphosphate triphosphohydrolase (dNTPase) activity of the EF1143 protein from Enterococcus faecalis and crystal structure of the activator-substrate complex. J Biol Chem 286:33158–33166. https://doi.org/10.1074/jbc.M111.250456
Czernecki D, Legrand P, Tekpinar M et al (2021) How cyanophage S-2L rejects adenine and incorporates 2-aminoadenine to saturate hydrogen bonding in its DNA. Nat Commun 12:2420. https://doi.org/10.1038/s41467-021-22626-x
Moroz OV, Harkiolaki M, Galperin MY et al (2004) The crystal structure of a complex of Campylobacter jejuni dUTPase with substrate analogue sheds light on the mechanism and suggests the “basic module” for dimeric d(C/U)TPases. J Mol Biol 342:1583–1597. https://doi.org/10.1016/j.jmb.2004.07.050
Czernecki D, Bonhomme F, Kaminski P-A, Delarue M (2021) Characterization of a triad of genes in cyanophage S-2L sufficient to replace adenine by 2-aminoadenine in bacterial DNA. Nat Commun 12:4710. https://doi.org/10.1038/s41467-021-25064-x
Cleaves HJ, Butch C, Burger PB et al (2019) One among millions: the chemical space of nucleic acid-like molecules. J Chem Inf Model 59:4266–4277. https://doi.org/10.1021/acs.jcim.9b00632
Acknowledgements
I thank Professor Patrick Trieu-Cuot for his support.
Funding
This study was funded from the French government’s Investissement d’Avenir program, Laboratoire d’Excellence Integrative Biology of Emerging Infectious Diseases (Grant No. ANR-10-LABX-62-IBEID).
Author information
Authors and Affiliations
Contributions
Not applicable.
Corresponding author
Ethics declarations
Conflict of interests
The author declares no competing financial interests.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Yes.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kaminski, P.A. Mechanisms supporting aminoadenine-based viral DNA genomes. Cell. Mol. Life Sci. 79, 51 (2022). https://doi.org/10.1007/s00018-021-04055-7
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-021-04055-7